Abstract
Background
Measures of malaria burden using microscopy and rapid diagnostic tests (RDTs) in cross-sectional household surveys may incompletely describe the burden of malaria in low-transmission settings. This study describes the pattern of malaria transmission in Ethiopia using serological antibody estimates derived from a nationwide household survey completed in 2015.
Methods
Dried blood spot (DBS) samples were collected during the Ethiopian Malaria Indicator Survey in 2015 from malarious areas across Ethiopia. Samples were analysed using bead-based multiplex assays for IgG antibodies for six Plasmodium antigens: four human malaria species-specific merozoite surface protein-1 19kD antigens (MSP-1) and Apical Membrane Antigen-1 (AMA-1) for Plasmodium falciparum and Plasmodium vivax. Seroprevalence was estimated by age, elevation and region. The seroconversion rate was estimated using a reversible catalytic model fitted with maximum likelihood methods.
Results
Of the 10,278 DBS samples available, 93.6% (9622/10,278) had valid serological results. The mean age of participants was 15.8 years and 53.3% were female. National seroprevalence for antibodies to P. falciparum was 32.1% (95% confidence interval (CI) 29.8–34.4) and 25.0% (95% CI 22.7–27.3) to P. vivax. Estimated seroprevalences for Plasmodium malariae and Plasmodium ovale were 8.6% (95% CI 7.6–9.7) and 3.1% (95% CI 2.5–3.8), respectively. For P. falciparum seroprevalence estimates were significantly higher at lower elevations (< 2000 m) compared to higher (2000–2500 m) (aOR 4.4; p < 0.01). Among regions, P. falciparum seroprevalence ranged from 11.0% (95% CI 8.8–13.7) in Somali to 65.0% (95% CI 58.0–71.4) in Gambela Region and for P. vivax from 4.0% (95% CI 2.6–6.2) in Somali to 36.7% (95% CI 30.0–44.1) in Amhara Region. Models fitted to measure seroconversion rates showed variation nationally and by elevation, region, antigen type, and within species.
Conclusion
Using multiplex serology assays, this study explored the cumulative malaria burden and regional dynamics of the four human malarias in Ethiopia. High malaria burden was observed in the northwest compared to the east. High transmission in the Gambela and Benishangul-Gumuz Regions and the neglected presence of P. malariae and P. ovale may require programmatic attention. The use of a multiplex assay for antibody detection in low transmission settings has the potential to act as a more sensitive biomarker.
Electronic supplementary material
The online version of this article (10.1186/s12936-019-2874-z) contains supplementary material, which is available to authorized users.
Keywords: Multiplex serology, Seroprevalence, Malaria, Ethiopia
Background
Malaria transmission in Ethiopia is non-stable, heterogeneous and seasonal, with peak periods following the main rains from June to September and short rains from March to April [1]. Historically, malaria epidemics occurred at 5 to 8-year intervals. The last malaria epidemic in 2004/2005 caused two million clinical cases and claimed approximately 3000 lives [2, 3]. Although malaria transmission has been declining over the past 15 years [4, 5], 60% of the Ethiopian landmass is suitable for malaria transmission and communities remain vulnerable for malaria transmission. The National Malaria Control Programme of Ethiopia considers areas below 2000 m elevation as malaria endemic and eligible for malaria control activities [6]. Plasmodium falciparum and Plasmodium vivax are the major documented malaria parasite species, with approximately 1.2 million P. falciparum cases and 678,000 P. vivax cases reported in 2015 [7]. The proportion of P. vivax cases has been increasing whereas a decline in P. falciparum incidence was observed [8].
With the decline in malaria cases, measuring malaria transmission intensity and dynamics is becoming increasingly challenging. Transmission estimates using standard malaria diagnostic tests, such as microscopy or rapid diagnostic tests (RDT), entomological inoculation rate, and splenomegaly prevalence fail to detect changes in the malaria burden in very low transmission settings [9]. The last three Ethiopian Malaria Indicator Surveys (EMIS) reported very low prevalence, using microscopy and RDTs: 0.9% by microscopy in 2007, 1.3% by microscopy and 4.5% by RDTs in 2011, and 0.5% by microscopy and 1.2% by RDT in 2015 [10–12]. With decreasing parasite prevalence as well as density of individual infections, the sensitivity of conventional diagnostic tools has been declining [13, 14]. Serological and molecular epidemiological studies may be more useful in such scenarios, and are being increasingly used for measuring malaria burden in low transmission settings [15–17]. Robust seroprevalence estimates of Plasmodium infections could be critical for monitoring and evaluation of ongoing malaria control and elimination activities in Ethiopia.
In this study, multiplex serological methods were used to estimate malaria exposure, transmission patterns, and spatial and regional distribution of malaria in Ethiopia, using samples collected during the 2015 EMIS.
Methods
Study area
The study was conducted as part of the National Ethiopian Malaria Indicator Survey in 2015 (EMIS-2015). Ethiopia has successfully implemented three national EMISs in 2007, 2011 and 2015 [10–12]. EMIS-2015 was embarked during the peak of malaria transmission season, between September and December 2015. The sampling design provided national and sub-national estimates of major malaria intervention indicators, including malaria prevalence for malarious areas of the country, as well as estimates for areas below 2000 m elevation, areas between 2000 and 2500 m, and for 10 administrative regions.
Study design and sample collection
EMIS-2015 was a cross-sectional survey that used a two-stage cluster sampling approach. A total of 555 enumeration areas (EAs) were selected proportional to the population size of the regions, as estimated by the Ethiopian Central Statistics Agency. In each EA, 25 households were randomly selected following onsite EA household mapping. A total of 53,335 individuals were surveyed in 13,875 selected households. Demographic, socio-economic, use of malaria prevention, and malariometric data were collected in the selected households from each EA. Children under 5 years of age in each selected household and persons of all ages in every fourth household were eligible for biological sample testing. Whole blood from a finger prick from consenting individuals was collected for a malaria RDT (histidine‐rich protein 2 (HRP2) and Plasmodium lactate dehydrogenase (pLDH) antigen CareStart®, AccessBio, US), a malaria blood slide (thick and thin blood film), haemoglobin (Hemocue Hb 201+ , Hemocue AB, Ängelholm, Sweden) measurement, and dried blood spot (DBS) samples. Whatman 903 Protein Saver (GE Healthcare, Pittsburgh, PA, USA) filter paper was used for DBS collection. Filter papers were air dried, individually packed in a plastic bag together with a desiccant and stored at − 20 °C at the Ethiopian Public Health Institute (EPHI) before they were shipped for further laboratory-based testing at the US Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA.
Ethical consideration
The EMIS-2015 protocol received ethical clearance from the National Research Ethics Review Committee of Ethiopia and PATH. The survey protocol underwent human subjects review at CDC and was considered to be a non-research programme evaluation activity. Additional ethical clearance for the present serology study was obtained from the Institutional Review Board of the College of Health Sciences of the Addis Ababa, University (AAUMF 03-008).
Blood spot elution, bead coupling and antigens used in the study
Laboratory analysis was conducted as previously described [15]. Briefly, a 6-mm punch was used from each DBS sample and blood eluted to a 1:20 concentration in blocking buffer (Buffer B: Phosphate Buffered Saline (PBS) containing 0.5% Bovine Serum Albumin (BSA), 0.05% Tween 20, 0.02% sodium azide, 0.5% polyvinyl alcohol, 0.8% polyvinylpyrrolidone and 0.5% w/v Escherichia coli extract) overnight.
Six Plasmodium antigens were used for this study: two P. falciparum antigens (Merozoite Surface Protein-1 19kD (MSP-1) and Apical Merozoite Antigen-1 (AMA-1), two P. vivax antigens (PvMSP-1 and PvAMA-1), one P. malariae (PmMSP-1) and one P. ovale (PoMSP-1). The four Plasmodium MSP-1 19kD antigens were produced as recombinant proteins and purified as described previously [18]. The external domain of P. falciparum AMA-1 antigen was produced at the Walter Reed Army Institute of Research (WRAIR) under previously published conditions [19]. The P. vivax AMA-1 antigen was produced at the London School of Hygiene and Tropical Medicine (LSHTM) under previously published conditions [20]. The Schistosoma japonicum glutathione-S-transferase antigen (GST) was produced recombinantly and served as a generic protein to assess immunoglobulin G (IgG) non-specific binding.
All antigens were coupled to magnetic beads (Luminex Corp, Austin, TX, USA) as per prior studies [18, 21]. Briefly, beads were pulse vortexed, transferred to a microcentrifuge tube and centrifuged for 1.5 min at 13,000g. Supernatant was removed, and the beads were washed with a 0.1 M and pH 6.2 sodium phosphate (NaP) solution. Beads were activated by suspending in NaP with 5 mg/mL of EDC (1-ethyl-3-[3-dimethylaminutesopropyl] carbodiimide hydrochloride) and 5 mg/mL sulfo-NHS (sulfo N-hydroxylsulfosuccinimide) and incubating with rotation for 20 min at room temperature (RT), while protected from light. After a wash with coupling buffer (50 mM 2-(4-morpholino)-ethane sulfonic acid, 0.85% NaCl at pH 5.0), antigens were coupled to beads in the presence of a coupling buffer for 2 h at a concentration of 20 ug/mL for all antigens, except for PvAMA1 and GST at 15 ug/mL. Beads were washed once with PBS and suspended in PBS with 1% bovine serum albumin (BSA) with incubation for 30 min at RT by rotation. Beads were then resuspended in a storage buffer (PBS, 1% BSA, 0.02% sodium azide and 0.05% Tween-20) and stored at 4 °C. Coupled beads were run with a panel of known malaria seronegatives to assure minimal non-specific MFI signal would be given by beads used in this study.
The DBS elution was assayed for IgG antibodies using bead-based multiplex technology, and all wash steps were performed with a handheld magnet. In 5 mL of reagent buffer (Buffer A: PBS, 0.5% BSA, 0.05% Tween-20, 0.02% NaN3), a bead mix was prepared with all coupled bead regions included (approximately 625 beads/antigen per well), and 50 uL bead mix was pipetted into each well of a BioPlex Pro plate (BioRad, Hercules, CA, USA). Beads were washed twice with 100 µL PBS with Tween (PBST), and 50 µL of the reagent mix [in 5 mL Buffer A: 1:500 anti-human IgG (Southen Biotech), 1:625 anti-human IgG4 (Southern Biotech), 1:200 streptavidin-PE (Invitrogen)] was added to each well. Negative control samples and a dilution curve of hyperimmune serum were added to each assay plate to monitor any change in control values over the course of the study. Samples of blood eluted from DBS were added to predefined wells (already containing beads and reagent mix) at 1:50 dilution. Plates were incubated overnight with gentle shaking at RT and protected from light. The next morning (approximately 16 h total incubation time), plates were washed three times, and beads were re-suspended with 100 µL PBS and read on a MAGPIX machine (Luminex Corp, Austin, TX, USA). Mean fluorescence intensity (MFI) signal was generated for a minimum of 50 beads/region, and the background (bg) MFI from wells incubated with Buffer B was subtracted from each sample to give a final value of MFI-bg. Minimal variation was seen in the assay signal for the background or positive hyperimmune serum curve values, so no plates were re-run for the study.
Statistical analysis
Data analysis was done using Stata 13 software (College Station, USA). Samples with GST MFI-bg reads above 1000 MFI value (non-specific binding) were excluded from the analysis (n = 678). To dichotomize seropositivity, log10-transformed MFI-bg values were fitted to a two-component Finite Mixture Model (FMM) by the FMM procedure with normal distribution and maximum likelihood estimation outputs. A seropositivity cutoff value was determined by the mean MFI-bg value of the first (assumed seronegative) component plus three standard deviations [18]. Overall, P. falciparum and P. vivax seropositivity were defined as an individual being positive for either or both of the MSP-1 and AMA-1 antigens for each species. Seropositivity estimates for P. malariae and P. ovale are presented using the seropositivity cut-off method outlined above. However, to minimize cross-reactivity between the species-specific MSP-1 antigens, the current study report an additional conservative two-stage approach that increase specificity, but decrease sensitivity of the specific antibodies to the targeted antigens. For this conservative approach for P. malariae and P. ovale, individual readings for PmMSP-1 or PoMSP-1 response first had to be above the MFI-bg cutoff as described above. Additionally, the PmMSP-1 and/or PoMSP-1 MFI-bg signal for that sample also needed to be above the PfMSP-1 MFI-bg signal for the same sample (ratio to PfMSP-1 greater than 1.0) to be considered PmMSP-1 and/or PoMSP-1 positive.
A reversible catalytic model was fitted to the dichotomized data using maximum likelihood methods to generate a seroconversion rate (SCR or λ) and a seroreversion rate (ρ). The model was used to generate age seroprevalence curves, from which a seroconversion rate (SCR) representing the force of infection for the community was calculated. Evidence for two forces of infection was investigated, if visual inspection of SCR curves indicated such a comparison and was guided by a profile likelihood plots to determine the most likely time (year) of change in transmission [16, 22]. Multiple logistic regression models were employed to determine odds ratios (OR); 95% confidence intervals (CI) for gender, age and elevations. Similar regression models were employed for P. falciparum and P. vivax seropositivity. Adjustments were made by region, elevation and age group.
Empirical bayesian kriging in ArcGIS version 10.5 (ESRI, CA, USA) was used to predict the spatial distribution of seroprevalence as a continuous surface of probability of being above a cut-off. This is an established method that uses a statistical model to predict and interpolate the spatial distribution of a variable from available data [23]. Maps were developed separately for MSP-1 and AMA-1 antigens for both P. falciparum and P. vivax for all individuals and for children under 5 years of age. Seroprevalence by woreda/district was compared with the 2015 annual parasite incidence (API) data. QGIS version 2.1.8 and ArcGIS version 10.5 (ESRI, Redlands, CA) were used to produce maps.
Sampling weights calculated during MIS 2015 were used to ensure the representativeness of the samples tested to the study population.
Results
Study population
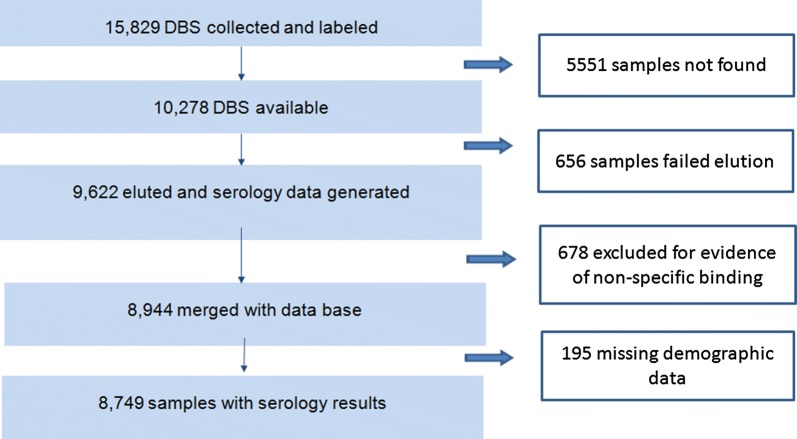
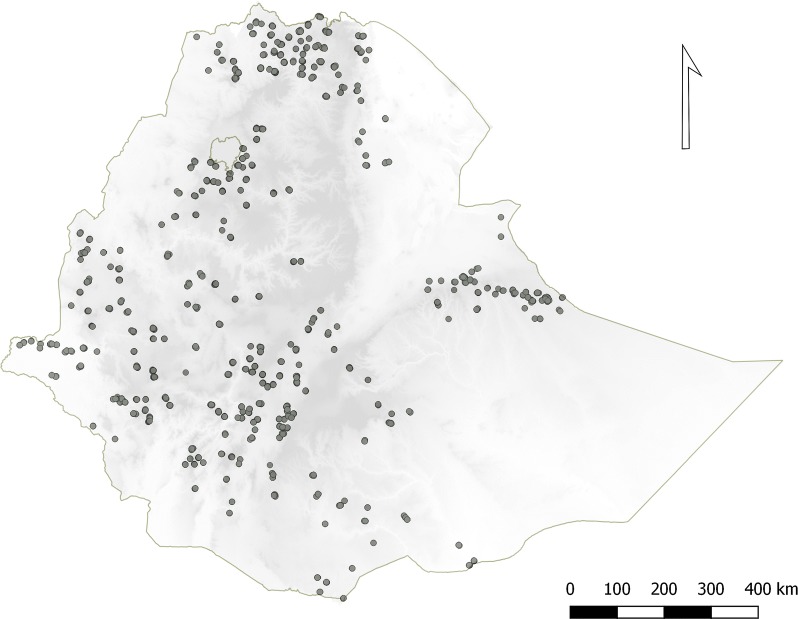
Of the expected 15,829 DBS samples, 10,278 individual DBSs were available for the current study, serological results were generated for 93.6% (9622/10,278) of these (Fig. 1). A total of 8944 results were merged with the EMIS-2015 data base, of which 52.3% (95% CI (50.8–53.7)) of the survey participants were female (Table 1). Figure 2 shows the spatial distribution of EAs providing serology data. The overall proportion of samples collected from children under 5 years was 44.8% (95% CI 43.0–46.5); although this proportion was above 60% in the Afar and Somali Regions (Table 1). The number of individuals with serology data within an EA ranged from 1 to 55, with a mean of 24 individuals per EA. Overall mean age was 15.8 years (95% CI 15.2–16.4) (range 1–107). Of the samples collected, 75.0% (95% CI 67.5–81.2) were from areas with elevation less than 2000 m.
Fig. 1.
Flow chart showing sample selection and analysis procedure
Table 1.
Characteristics of study participants by administrative region, Ethiopian Malaria Indicator Survey-2015 (EMIS-2015)
| Tigray | Afar | Amhara | Oromia | Somali | Benishangul-Gumuz | Southern Nations and Nationalities Peoples’ | Gambela | Harari | Diredawa | National | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender % (95% CI) | n = 8743 | ||||||||||
| Male | 46.5 (44.1–48.9) | 47.6 (42.9–52.3) | 50.9 (47.4–54.3) | 48.2 (45.6–50.9) | 45.1 (40.8–49.4) | 45.3 (40.8–49.9) | 46.4 (44.3–48.5) | 41.4 (37.4–45.5) | 47.4 (3.0–65.4) | 40.5 (33.1–48.4) | 47.7 (46.3–49.2) |
| Female | 53.3 (51.1–55.9) | 52.4 (47.7–57.1) | 49.1 (45.7–52.6) | 51.8 (49.1–54.4) | 54.9 (50.6–59.2) | 54.7 (50.1–59.2) | 53.6 (51.5–55.7) | 58.6 (54.5–62.6) | 52.6 (34.6–70.0) | 59.5 (51.6–66.9) | 52.3 (50.8–53.7) |
| Age group (years)% (95% CI) | |||||||||||
| 0–5 | 43.8 (39.6–48.0) | 66.6 (63.7–69.3) | 40.8 (37.1–44.7) | 44.6 (41.5–47.9) | 61.7 (58.8–64.6) | 49.5 (41.5–57.5) | 45.4 (43.2–47.7) | 54.8 (50.8–58.8) | 25.5 (12.9–44.2) | 46.0 (35.3–57.1) | 44.8 (43.0–46.5) |
| 5–15 | 17.7 (15.2–20.4) | 12.8 (9.2–17.5) | 20.5 (18.0–23.3) | 18.8 (15.8–22.3) | 14.6 (11.7–18.1) | 16.0 (12.9–19.6) | 20.8 (18.9–22.9) | 18.9 (16.6–21.5) | 3.5 (1.0–11.6) | 10.2 (6.6–15.4) | 19.4 (17.8–21.1) |
| 15–25 | 13.3 (11.3–15.6) | 6.1 (3.9–9.4) | 12.6 (9.5–16.6) | 11.6 (10.0–13.4) | 5.2 (3.6–7.5) | 11.5 (7.8–16.6) | 11.4 (9.7–13.4) | 10.7 (8.3–13.8) | 22.1 (17.4–27.5) | 14.0 (10.9–17.9) | 11.7 (10.7–12.8) |
| 25–50 | 17.1 (14.9–19.4) | 14.1 (12.1–16.4) | 19.2 (16.2–22.6) | 19.7 (17.5–22.1) | 14.6 (13.0–16.4) | 14.5 (10.7–19.3) | 17.1 (15.6–18.6) | 12.3 (1.1–15.0) | 36.8 (29.3–45.0) | 21.7 (15.9–28.9) | 18.5 (17.3–19.8) |
| 50+ | 8.2 (6.6–10.1) | 0.5 (0.1–1.5) | 6.9 (5.1–9.1) | 5.2 (3.9–7.0) | 3.8 (2.7–5.3) | 8.6 (5.3–13.5) | 5.2 (4.1–6.6) | 3.2 (2.3–4.5) | 12.1 (7.4–19.2) | 8.1 (4.5–14.2) | 5.7 (4.9–6.6) |
| Elevation (m) % (95% CI) | n = 8740* | ||||||||||
| < 1000 | 5.6 (1.6–17.4) | 38.0 (13.7–70.4) | 0 (0.0–0.0) | 0 (0.0–0.0) | 28.6 (12.6–52.7) | 8.2 (1.1–52.7) | 0 (0.0–0.0) | 88.4 (72.1–95.8 | 0 (0.0–0.0) | 0.9 (0.1–6.7) | 1.9 (1.2–2.8) |
| 1000–2000 | 64.3 (46.8–78.6) | 62.0 (29.6–86.3) | 53.4 (33.2–72.5) | 77.4 (47.3–87.4) | 71.4 (47.3–87.4) | 91.8 (57.4–98.9) | 79.0 (67.3–87.3) | 11.6 (4.2–27.9) | 100 (100.0–100.0) | 99.1 (93.9–99.9) | 73.1 (65.6–79.4) |
| > 2000 | 30.2 (16.5–48.5) | 0 (0.0–0.0) | 46.6 (27.5–66.8) | 22.6 (13.1–36.1) | 0 (0.0–0.0) | 0 (0.0–0.0) | 21 (12.7–32.7) | 0 (0.0–0.0) | 0 (0.0–0.0) | 0 (0.0–0.0) | 25.0 (18.8–32.5) |
| Bed net use (last night) % (95% CI) | n = 5168** | ||||||||||
| Yes | 57.6 (50.3–64.5) | 80.5 (65.7–89.9) | 59.1 (48.1–69.4) | 69.6 (59.6–78.1) | 75.2 (61.2–85.4) | 66.1 (54.3–76.2) | 55.7 (49.4–61.8) | 84.7 (76.0–90.6) | 33.5 (12.8–63.3) | 25 (14.3–41.6) | 62.1 (57.5–66.6) |
| No | 42.4 (35.5–49.7) | 19.5 (10.1–34.3) | 40.9 (30.6–51.9) | 30.4 (21.9–40.4) | 24.8 (14.6–38.8) | 33.9 (23.8–45.7) | 44.3 (38.2–50.6) | 15.3 (9.4–24.0) | 66.5 (36.7–87.2) | 74.4 (58.4–85.7) | 37.9 (33.4–42.5) |
| RDT results % (95% CI) | |||||||||||
| Pf | 0.8 (0.4–1.6) | 0 (0.0–0.0) | 0.7 (0.2–1.8) | 0.2 (0.1–0.6) | 0 (0.0–0.0) | 6.2 (2.2–16.3) | 0.2 (0.1–0.5) | 16.8 (11.8–23.2) | 0 (0.0–0.0) | 0 (0.0–0.0) | 0.5 (0.3–0.7) |
| Pan | 0.7 (0.3–1.3) | 0.2 (0.0–1.3) | 0 (0.0–0.0) | 0.1 (0.0–0.4) | 0.1 (0.0–0.8) | 0 (0.0–0.0) | 0.4 (0.1–1.2) | 0.1 (0.0–0.6) | 0 (0.0–0.0) | 0 (0.0–0.0) | 0.2 (0.1–0.4) |
| Pf/Pan | 0.1 (0.0–0.6) | 0 (0.0–0.0) | 0.5 (0.1–1.8) | 0 (0.0–0.0) | 0 (0.0–0.0) | 0.4 (0.0–2.6) | 0.2 (0.0–0.6) | 1.7 (0.9–3.0) | 0 (0.0–0.0) | 0 (0.0–0.0) | 0.1 (0.1–0.3) |
| Negative | 98.4 (97.4–99.0) | 99.8 (98.7–100.0) | 98.9 (96.7–99.6) | 99.7 (99.3–99.8) | 99.9 (99.2–100.0) | 93.5 (82.3–97.8) | 99.3 (98.3–99.7) | 81.5 (74.5–86.9) | 100 (100.0–100.0) | 100 100.0–100.0) | 99.2 (98.8–99.4) |
n, number of observations; RDT, rapid diagnostic test; *3 people did not have GPS coordinates; **3613 individuals were without bed net use data; ***RDT test was not done for 4 individuals; 4 RDT tests were invalid and 3 individuals refused RDT testing
Fig. 2.
Distribution of dried blood spot (DBS) samples collected from selected malaria endemic areas of Ethiopia as part of the national, cross-sectional, household Malaria Indicator Survey conducted in 2015 (EMIS-2015). Each point depicts a cluster of sampled households in an EA. Shading indicates elevation which ranges from − 131 m below sea level (lightest) to 4092 m (darkest)
A total of 0.8% (95% CI 0.6–1.2) of the participants were positive by RDT; comprising 0.5% (95% CI 0.3–0.7) for P. falciparum (HRP2 antigen positive), 0.2% (95% CI 0.1–0.4) for either P. vivax, P. malariae or P. ovale (Pv/Pm/Po) (pan-LDH antigen positive), and 0.1% (95% CI 0.1–0.3) considered as a mixed infection (HRP2 and pan-LDH positive). More than 90% of the positive P. falciparum RDT results were from the Gambela and Benishangual-Gumuz Regions. 62.1% (95% CI 57.5–66.6) of respondents said they used a bed net during the previous night (Table 1).
Seroprevalence
Nationally, 32.1% (95% CI 29.8–34.4) of the participants were seropositive for any IgG against the two P. falciparum antigens, and 25.0% (95% CI 22.7–27.34) for P. vivax. Only 6.6% (95% CI 5.5–8.0) of participants were seropositive for both P. falciparum and P. vivax, while 41.1% (95% CI 38.5–43.7) of individuals were positive either for P. falciparum or P. vivax. Eight point six percent (95% CI 7.6–9.7) and 3.1% (95% CI 2.5–3.8) were seropositive for P. malariae and P. ovale, respectively (Table 2). By antigen, 23.8% (95% CI 21.4–26.3) were positive against P. falciparum MSP-1 and 24.5% (95% CI 22.4–26.7) AMA-1, while for P. vivax, 21.2% (95% CI 19.2–23.4) were positive against MSP-1 and 14.4% (95% CI 12.7–16.2) AMA-1.
Table 2.
Proportion of individuals (all ages and under 5 years) found to be malaria seropositive by species and by antigen in malaria endemic areas of Ethiopia, 2015
| Plasmodium species | Antigens | % seropositive (95% CI) (all ages) | % seropositive (95% CI) (under 5 years) |
|---|---|---|---|
| P. falciparum | MSP-1 | 23.8 (21.4–26.3) | 12.3 (10.6–14.2) |
| AMA-1 | 24.5 (22.4–26.7) | 11.3 (9.8–13.0) | |
| Pf positive (MSP-1 and/or AMA-1) | 32.1 (29.8–34.4) | 18.0 (16.1–20.1) | |
| P. vivax | MSP-1 | 21.2 (19.2–23.4) | 12.3 (10.5–14.4) |
| AMA-1 | 14.4 (12.7–16.2) | 6.7 (5.5–8.0) | |
| Pv positive (MSP-1 and/or AMA-1) | 25.0 (22.7–27.3) | 14.8 (12.7–17.2) | |
| P. malariaea | MSP-1 | 8.6 (7.6–9.7) | 6.1 (5.0–7.3) |
| P. ovale a | MSP-1 | 3.1 (2.5–3.8) | 2.0 (1.4–2.7) |
| Any Plasmodium | Pf (MSP-1/AMA-1) or Pv (MSP-1/AMA-1) or Pm MSP-1 or Po MSP-1 | 44.5 (41.9–47.2) | 30.2 (27.3–33.2) |
| Either P. falciparum or P. vivax | Pf (MSP-1/AMA-1) or Pv (MSP-1/AMA-1) | 41.1 (38.5–43.7) | 26.7 (24.0–29.5) |
| Both P. falciparum and P. vivax | Pf (MSP-1/AMA-1) and Pv (MSP-1/AMA-1) | 6.6 (5.5–8.0) | 1.7 (1.2–2.4) |
Samples positive for Merozoite Surface Protein-1 (MSP-1) and/or Apical Membrane Antigen-1 (AMA-1) for the respective species were considered positive for that species
Pf, Plasmodium falciparum; Pm, Plasmodium malariae; Po, Plasmodium ovale; Pv, Plasmodium vivax
aNormal approach (mean seronegative +3sd) to defining cut-off level used for P. malariae and P. ovale
For children under 5 years of age, 18.0% (95% CI 16.1–20.1) and 14.8% (95% CI 12.7–17.2) were seropositive for P. falciparum and P. vivax, respectively. Similarly, 33.3% (95% CI 30.4–36.2) of male children were seropositive for P. falciparum compared to 30.9% (95% CI 28.3–33.6) of the females [aOR: 1.3 (95% CI 1.1–1.4), p < 0.05] (Additional file 1). A similar trend was observed for P. vivax, where 26.4% (95% CI 23.4–29.7) of males compared to 23.6% (95% CI 21.1–26.3) of females were seropositive [aOR: 1.3 (95% CI 1.1–1.5), p > 0.05].
Seroprevalence by age
The DBS sample collection was purposely biased towards children under 5 years of age, with 44.8% (95% CI 43.0–46.5) of the DBS samples having been collected from this age group. Seropositivity increased with age group. This pattern was more pronounced for P. falciparum than P. vivax, although the difference was significant in both Plasmodium species (p < 0.05).
Conservative estimate for Plasmodium malariae and Plasmodium ovale
Table 3 shows of all samples tested, 5.1% (95% CI 4.4–6.0) were seropositive for P. malariae and 1.5% (95% CI 1.1–2.0) for P. ovale using the conservative approach to define seropositivity. Both species were observed in all the regions, albeit with a variable distribution. The proportion of seropositives was slightly lower for both species, accordingly to the conservative estimation method. A similar trend of seropositivity was observed for children under 5 years of age (Table 3).
Table 3.
Proportion seropositive for less common malarias [P. malariae Merozoite Surface Protein-1 (PmMSP-1) and P. ovale Merozoite Surface Protein-1 (PoMSP-1)], using the conservative approach to defining seropositive cut-off values, by region and age group, in malaria endemic areas of Ethiopia, 2015
| Region | All ages | Under 5 years | ||
|---|---|---|---|---|
| PmMSP-1 | PoMSP-1 | PmMSP-1 | PoMSP-1 | |
| Tigray | 1.1 (0.6–2.3) | 0.6 (0.3–1.1) | 1.1 (0.4–2.9) | 0.3 (0.1–1.2) |
| Afar | 1.3 (0.6–3.0) | 0.6 (0.1–2.7) | 1.8 (0.7–4.2) | 0.5 (0.1–4.2) |
| Amhara | 5.0 (3.2–7.6) | 2.4 (1.8–3.3) | 2.7 (1.1–6.2) | 1.0 (0.3–3.1) |
| Oromia | 6.3 (5.0–7.8) | 2.0 (1.3–3.0) | 5.8 (4.4–7.8) | 1.8 (1.0–3.3) |
| Somali | 1.3 (0.6–2.8) | 0.1 (0.0–0.8) | 1.1 (0.4–2.9) | 0 |
| Benishangul-Gumuz | 2.1 (1.0–4.4) | 0.7 (0.2–2.5) | 1.3 (0.4–4.7) | 1.5 (0.4–4.9) |
| Southern Nation and Nationalities People’s | 4.9 (3.7–6.3) | 0.6 (0.4–1.2) | 4.7 (3.1–7.1) | 0.7 (0.3–1.8) |
| Gambela | 0.2 (0.1–1.1) | 0.1 (0.0–0.5) | 0.3 (0.1–1.2) | 0 |
| Harari | 4.5 (0.6–26.4) | 4.0 (0.3–33.6) | 0 | 0 |
| Dire Dawa | 2.4 (0.7–8.0) | 0.8 (0.1–5.3) | 1.7 (0.3–9.9) | 0 |
| Total (%) | 5.1 (4.4–6.0) | 1.5 (1.1–2.0) | 4.6 (3.6–5.7) | 1.2 (0.8–1.9) |
Pm, Plasmodium malariae; Po, Plasmodium ovale
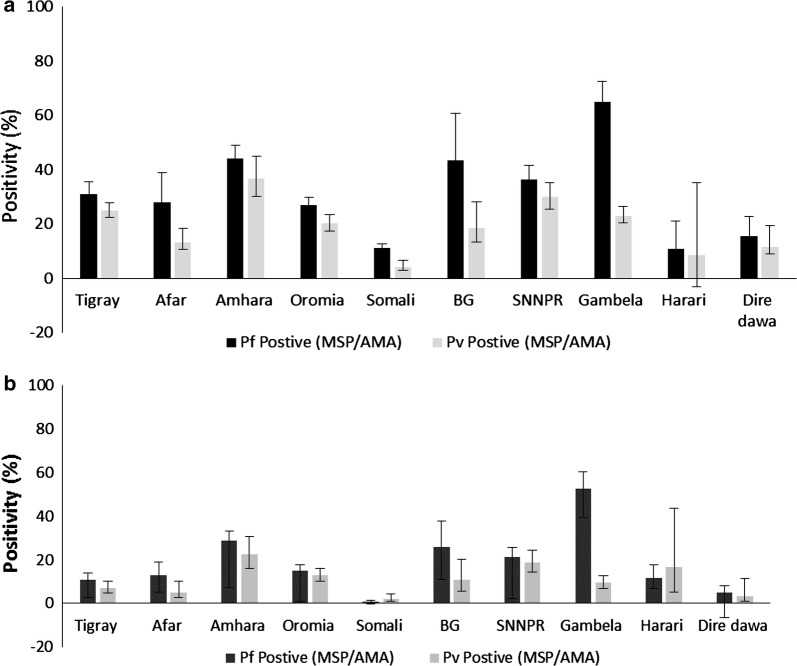
Seroprevalence by region
The proportion of individuals seropositive was variable by region, with P. falciparum seropositivity higher than P. vivax seropositivity in all the regions (Fig. 3). The proportion seropositive for P. falciparum by region ranged from 11.0% (95% CI: 8.8–13.7) in Somali to 65.0% (95% CI: 58.0–71.4) in Gambela Region and for P. vivax from 4.0% (95% CI: 2.6–6.2) in Somali to 36.7% (95% CI: 30.0–44.1) in Amhara Region.
Fig. 3.
Mean seropositivity by region for Plasmodium falciparum and Plasmodium vivax, 2015. An individual was considered positive if they had a positive antibody response either for Merozoite Surface Protein-1 (MSP-1) or Apical Membrane Antigen-1 (AMA-1) antigens for each species. a All ages; b children under 5 years of age. Error bars indicate the 95% confidence interval (BG: Benishangul-Gumuz and SNNPR: Southern Nations and Nationalities People’s Region)
The API data from 2014 (API 2014) obtained from the Federal Ministry of Health (Additional file 2), observed high malaria transmission in Gambela, Benishangul-Gumuz and Amhara Regions. By antigen comparison, P. falciparum API showed greater association with PfMSP-1 [Rho of 0.37 and p < 0.01 (spearman correlation)] and P. vivax API with PvAMA-1 (Rho 0.32 and p < 0.01).
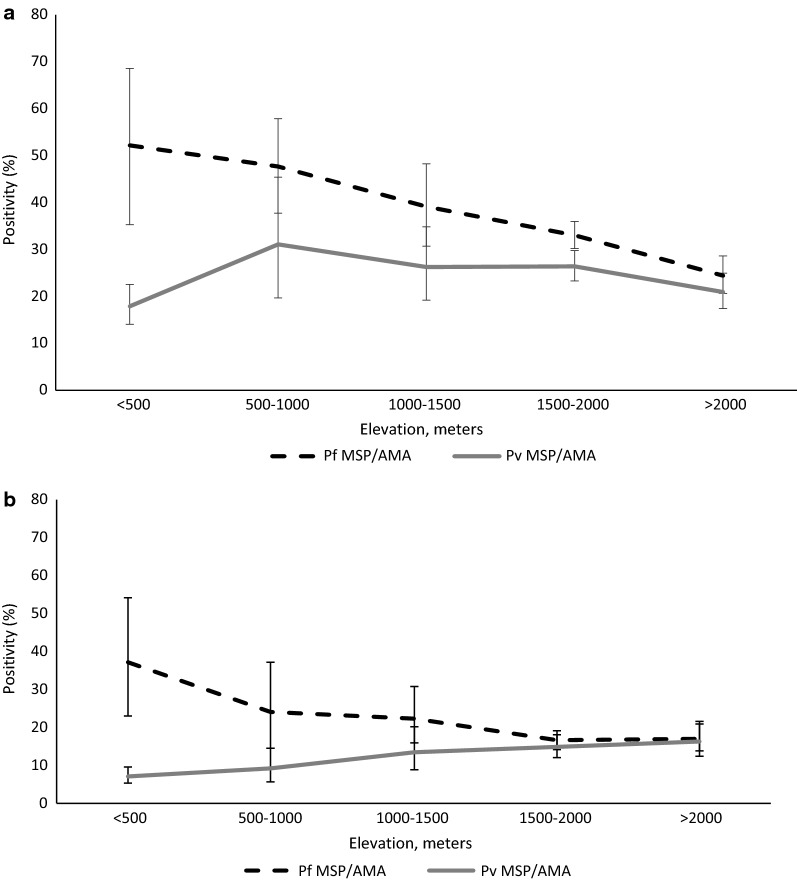
Seroprevalence by elevation
Seropositivity was inversely related to elevation for P. falciparum, with lower elevation (< 2000 m) areas having significantly higher seroprevalence compared to areas of higher elevation (2000–2500 m) [aOR 4.36 (95% CI 2.7–7.0), p < 0.01]. The highest seropositivity was observed for P. falciparum at elevations below 1500 m and a peak was observed at an elevation below 1000 m (Fig. 4a). A weaker inverse effect of elevation was observed for P. vivax. There was no significant difference in the transmission of P. vivax between above and below 2000 m (OR 1.5 (95% CI 0.9–2.4), p > 0.05). Among children under 5 years of age, P. falciparum and P. vivax both showed a significant although varying relationship with elevation. Plasmodium vivax showed direct relation with increasing elevation. High P. vivax seropositivity was observed at higher elevations above 1500 m (Fig. 4b). No significant effect of elevation on seropositivity was detected for the rare species, P. malariae and P. ovale (p > 0.05).
Fig. 4.
Mean seropositivity for Plasmodium falciparum Merozoite Surface Protien-1 (MSP-1) or Apical membrane Antigen-1 (AMA-1), and Plasmodium vivax MSP-1 or AMA-1 by elevation category, 2015. An individual was considered positive if they had a positive antibody response either for Merozoite Surface Protien-1 (MSP-1) or Apical Membrane Antigen-1 (AMA-1) antigens for each species. Broken line depicts P. falciparum (MSP-1/AMA-1) and solid line represent P. vivax (MSP-1/AMA-1). a All ages; b children under 5 years of age. Error bars indicate the 95% confidence interval
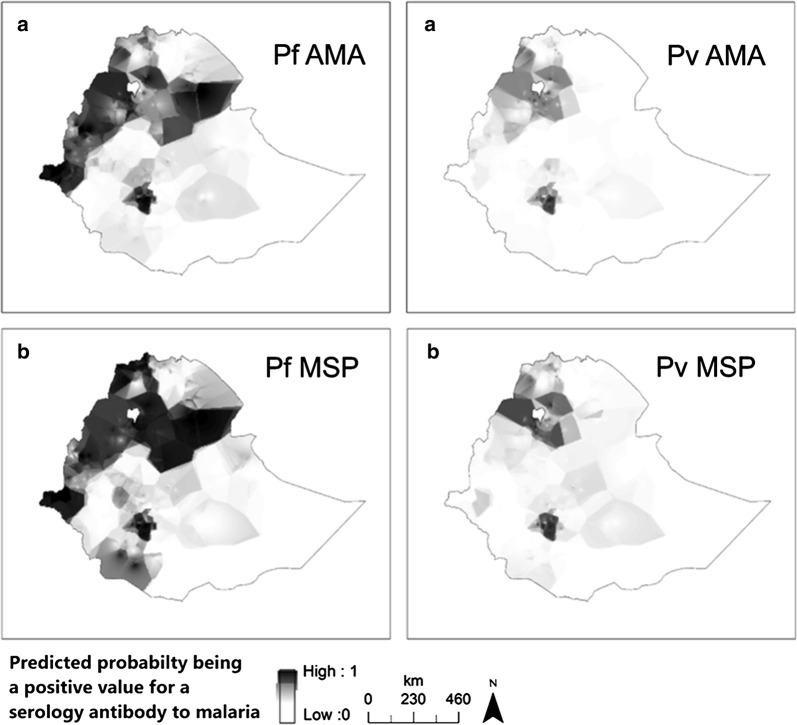
Generally, seropositivity was found to vary in space, with elevation and among administrative regions, as depicted in Fig. 5. An interpolated surface map predicted differing transmission patterns for P. falciparum and P. vivax. Higher seropositivity is predicted in the northwest compared to the southeast.
Fig. 5.
An interpolated surface map for Plasmodium falciparum antigen responses [a Apical Membrane Antigen-1 (AMA-1) and b Merozoite Surface Protein-1 (MSP-1)] and Plasmodium vivax antigen responses (a AMA-1 and b MSP-1). Darker colour shows high probability of being above the cut-off for seropositivity and lighter colour lower. Empirical bayesian kriging was used to produce the maps from serology data from samples collected during Ethiopian Malaria Indicator Survey (EMIS 2015)
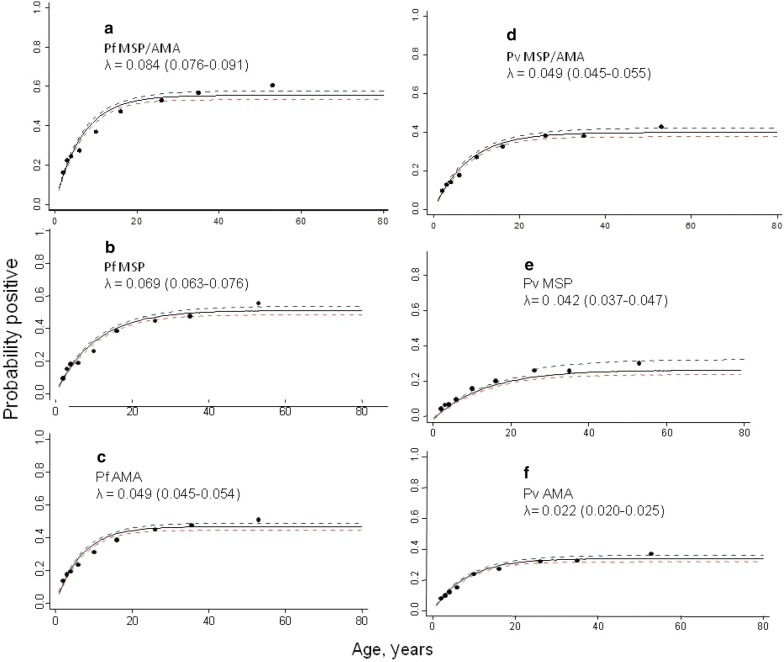
Seroconversion curves
The overall age seroconversion plot for antibody responses for both P. falciparum and P. vivax are shown in Fig. 6. In general, higher seroconversion rates were observed for P. falciparum compared to P. vivax both for the combined MSP-1/AMA-1 outcome and individual MSP-1 and AMA-1 antigens; and MSP-1 antigen show higher seroconversion rates compared to AMA-1 antigens for both species. Heterogeneity was observed across the different antigen responses. A similar trend was observed when the model was fitted to data with elevation below 2000 m and combined MSP-1 and AMA-1 antigens. Regionally, the seroconversion model was fitted to the four major administrative regions (Amhara, Oromia, SNNP, Tigray) both for P. falciparum and P. vivax MSP-1 and AMA-1 antigen responses. Heterogeneous seroconversion rates were observed over the regions (Additional files 3 and 4). For seroconversion plots, where visual inspection indicated better fit between the observed and predicted pattern of transmission, two forces of infection were compared with a single force of infection. This showed a non-significant change over around two decades for both P. falciparum and P. vivax antigen responses (p > 0.05).
Fig. 6.
National age-seroconversion plots for antibody responses for Plasmodium falciparum and Plasmodium vivax antigens [Merozoite Surface Protien-1 (MSP-1) and Apical Membrane Antigen-1 (AMA-1)]. Y-axis represents probability of being seropositive and the X-axis age. Seroconversion curves represent the rate at which a population becomes seropositive to specific antigens resulting in seroconversion rates (SCR) or lambda (λ). In each graph points represent age seroprevalence (by deciles), unbroken line represents maximum likelihood curves and broken lines represent the 95% confidence interval. a and d depict the seroconversion rates for combined (MSP-1/AMA-1) P. falciparum antigen response (a) and for combined (MSP-1/AMA-1) P. vivax antigen response (d). Plots b and c show seroconversion rates for individual P. falciparum antigens MSP-1 (b) and AMA-1 (c). Plots e and f show seroconversion rates for individual P. vivax antigens MSP-1 (e) and AMA-1 (f)
Discussion
The current study documented malaria seroprevalence in Ethiopia using samples collected from a national EMIS conducted in 2015. Of all samples analysed, 32.1% had IgG antibodies against combined P. falciparum MSP-1 or AMA-1 antigens, and 25.0% for P. vivax MSP-1 or AMA-1. The relative numbers were lower for children under 5 years of age at 18.0% for P. falciparum and 14.8% for P. vivax, suggesting a lower recent transmission in Ethiopia. Seropositivity in children under 5 years of age may accurately represent recent infection as they have had limited exposure and time to generate the cumulative antibody response seen in adults [24–26]. The serological results were cumulative antibody responses for the relatively long-term markers of transmission, MSP-1 and AMA-1 antigens [16, 22, 27–29]. The RDT results for the study samples showed 0.8% positivity for Plasmodium, of which 0.5% was due to P. falciparum.
Generally, significantly higher levels of P. falciparum transmission were observed in areas of lower elevation (< 2000 m). This is consistent with previous studies [1, 17, 19], and attributed to the ambient temperature and moisture requirements of the Anopheles vector mosquitoes [30]. This reaffirms the national malaria control programme’s strategy in which high elevation areas (> 2000 m) are considered to be areas where malaria is not transmitted and thus not targeted for vector control but only case management interventions. However, the evidence of increasing P. vivax seropositivity with elevation over the range of the study setting could expand the understanding of malaria transmission in Ethiopia. The relapse of P. vivax infections may have contributed to the high P. vivax prevalence observed at higher elevations [31–35]. Travel history is reported to be a risk factor for malaria exposure in Ethiopia [24–26], but in the EMIS-2015 only 2% reported travel history a month prior to the survey [12].
Seropositivity was high for children under 5 years of age, particularly in the Gambela and Benishangul Gumuz Regions, although was lower than in adults. These show higher malaria transmission that may need a more intensive approach for malaria control in the affected areas. A larger proportion of males than females were seropositive for both P. falciparum and P. vivax. Increased malaria risk in men in Ethiopia has been attributed to a greater extent of outdoor, occupational exposure [24–27, 36].
A total of 8.6% of individuals were seropositive for P. malariae and 3.1% for P. ovale. Variable trends of proportion seropositive were observed in the regions for both species. These findings, albeit with high proportion, support previous reports of sporadic occurrence of P. malariae and P. ovale that may be responsible for up to 1% of the malaria incident cases in Ethiopia [30, 37–40]. Currently there is no reporting system for these Plasmodium species, P. malariae and P. ovale, in Ethiopia. Antibody cross-binding among the different MSP-1 antigens has been documented as non-substantial [18, 41]. In the present study, data was analysed using an additional conservative approach to defining seropositivity to rule out the possibility of IgG cross-binding, thus reporting the most conservative estimates for comparison. Using this approach would potentially increase specificity but decrease sensitivity. The overall seropositivity, although reduced, was 5.1% for P. malariae and 1.5% for P. ovale, with the regional trend and pattern of transmission remaining similar.
The use of serological methods in measuring malaria transmission in low-transmission settings has been documented in a number of studies [15, 16, 27, 36, 42, 43]. Application of serology methods was suggested for Ethiopia because of the very low prevalence of malaria through RDT and microscopy methods across several national surveys [27, 36, 42]. The estimate was too low to track progress over the years or give detailed information about differences in malaria burden by region or risks of infection due to demographic or behavioural factors. By describing exposure to malaria over a longer period, serology data were able to better document differences in exposure between age groups, regions and by elevation.
The findings show the magnitude of transmission intensity was highly variable among different regions and elevations in Ethiopia. Yalew et al. reported similar trends with 30.0 and 21.8% seroprevalence for P. falciparum and P. vivax, respectively, for the Amhara region in 2013 [36]. The current study reported 32.1% for P. falciparum and 25.0% for P. vivax for the same region. The slight variation in seropositivity may be attributed to the different type of laboratory assay used. The challenge of comparing data from ELISA and bead-based multiplex serology has been investigated previously [15]. Results from multiplex serology were more robust, sensitive and ideal for large-scale surveys compared to ELISA [15]. Heterogeneity of response for the different antigens was demonstrated in several studies. Ashton et al. [27] described MSP-1 antigens to be better biomarkers for P. falciparum, although Cook et al. [16] reported AMA-1 to be better for measuring P. falciparum in low-transmission settings. Characterization of antigen markers may be required to further qualify the serology methods [44]. The current finding was in agreement with the API data from 2014 obtained through routine surveillance. Although routine surveillance can provide crucially important data for decision making for countries at lower transmission, additional biomarkers to track malaria burden can serve as ancillary data as surveillance systems continue to improve and dynamically evolve in completeness and quality.
The study had several limitations. Only 65% of DBS samples from the EMIS-2015 survey were available for serological analysis. Although no systematic bias was introduced and frequencies were weighted, the representativeness of this sample at the national and regional levels should be interpreted cautiously. The data were insufficient to produce refined spatial heat maps and the maps produced show only general trends. Samples collected are assumed to reflect the malaria exposure of their respective locations and did not account for potential population movement. Although the survey reported very low recent travel history, past travel encompassing the previous harvest season as well as previous years were not captured and could reflect past malaria exposure risk from a different locale. This study assessed long-term markers of transmission, but further characterizing antigens based on local eco-epidemiology is recommended to identify and select specific short- and long-term markers.
Conclusion
The current study explored the malaria burden and transmission intensity using serological markers of exposure to four malaria species. It documented the magnitude, spatial distribution and dynamics of transmission over differing elevation, administrative regions, and age groups using samples collected during a national programme evaluation survey with minimal sample preparation time and cost. These findings confirmed the National Malaria Control Programme data of the relative high transmission in the Gambela and Benishangul-Gumuz Regions and highlighted gaps in the current understanding of malaria epidemiology. The increase of P. vivax seropositivity with elevation and the presence of P. malariae and P. ovale in all administrative regions (given P. ovale requires hypnozoiticidal treatment) may require programmatic consideration in selecting diagnostic tools and appropriate treatment guidelines. These results can be used as baseline data to evaluate the recent malaria elimination efforts in Ethiopia especially as routine surveillance systems are progressively being strengthened.
Additional files
Additional file 1. Percent malaria seropositivity and Odds Ratio (OR) by sex, age and elevation for each species in malaria endemic areas of Ethiopia, 2014.
Additional file 2. An Interpolated surface map for P. falciparum (left) and P. vivax (right). Empirical bayesian kriging was used to produce the maps from the Annual Parasite Incidence data in Ethiopia (API 2014).
Additional file 3. Regional age-seroconversion plots for antibody responses for Plasmodium falciparum antigens. Y-axis represents probability of being seropositive and the X-axis age. Seroconversion curves represent the rate at which a population become seropositive to specific antigens resulting in seroconversion rates (SCR) or lambda (λ). In each graph points represent age seroprevalence (by deciles), unbroken line represents maximum likelihood curves and broken lines represent the 95% confidence interval. Plots A and B depict the seroconversion curves for P. falciparum antigens response to MSP-1 (A) and AMA-1(B) for the major four regions Tigray, Amhara, Oromia and Southern Nations and Nationalities People’s Regions.
Additional file 4. Regional age-seroconversion plots for antibody responses for P. falciparum antigens. Y-axis represents probability of being seropositive and the X-axis age. Seroconversion curves represent the rate at which a population become seropositive to specific antigens resulting in seroconversion rates (SCR) or lambda (λ). In each graph points represent age seroprevalence (by deciles), unbroken line represents maximum likelihood curves and broken lines represent the 95% confidence interval. Plots A and B depict the seroconversion curves for P. vivax antigens response to MSP-1 (A) and AMA-1 (B) for the major four regions, Tigray, Amhara, Oromia and Southern Nations and Nationalities People’s Region (SNNPR).
Acknowledgements
We acknowledge the MIS 2015 steering committee, data collectors and study participants for the samples collected. We thank Zewditu Bekele, Fitsum Tesfaye, Hilina Legesse and Hailemariam Difabachew from the Ethiopian Public Health Institute for sample organization and management.
Abbreviations
- AMA-1
Apical Membrane Antigen-1
- API
annual parasite incidence
- aOR
adjusted odds ratio
- BG
Benishangul-Gumuz
- BSA
bovine serum albumin
- CDC
Center for Disease Control
- CI
confidence interval
- DBS
dried blood spot
- EA
enumeration area
- ELISA
Enzyme Linked Immunosorbent Assay
- EMIS
Ethiopia Malaria Indicator Survey
- FMM
Finite Mixture Model
- GST
Gluthathione-S-Transferase
- HRP2
Histidin Rich Protein 2
- IgG
Immune globulin G
- LSHTM
London School of Hygiene and Tropical Medicine
- MFI
mean fluorescence intensity
- MSP-1
Merozoite Surface Protein-1
- RERRC
National Ethics and Research Review Committee
- OR
odds ratio
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- pLDH
plasmodium lactate dehydrogenase
- RDT
rapid diagnostic test
- RFLP
restriction fragment length polymorphism
- RT
room temperature
- SCR
seroconversion rate
- SNNPR
Southern Nations and Nationalities People’s Region
- WRAIR
Walter Reed Army Institute of Research
Authors’ contributions
Conceived the study: AA, JH, AAA, WD designed the experiments: AA, ER, YPS, BW. Performed the experiments: AA, ER, KG, BM. Analysed the data: AA, ER, JK, CD, RA, RM, IS, JH. Assisted study implementation: AAA, WD, AK, HS, JM, HM, JH. Critically reviewed and contributed to writing: JH, LS, ER, AAA GW, RM, WD. Drafted the manuscript: AA. All authors read and approved the final manuscript.
Funding
This research was made possible through support provided by the Global Fund to fight AIDS, Tuberculosis and Malaria via the Federal Ministry of Health in Ethiopia and the US President’s Malaria Initiative via the Office of Health, Infectious Diseases, and Nutrition, Bureau for Global Health, US Agency for International Development. The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of any collaborating institute including the Centers for Disease Control and Prevention or the US Agency for International Development. HT and JH receive salary support from the US President’s Malaria Initiative.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The MIS 2015 protocol received ethical clearance from the National Ethics and Research Review Committee (RERRC) of Ethiopia and from the College of Health Science Addis Ababa, University. The survey protocol which included serology testing underwent human subjects review at CDC and received non-research determination.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zein A, Koos H. The ecology of health and disease in Ethiopia. Addis Ababa: Ministry of Health; 1993. [Google Scholar]
- 2.Ethiopian Public Health Association (EPHA). Proceeding on the 2003 malaria epidemics in Ethiopia. EPHA Addis Ababa 2004.
- 3.Checchi F, Cox J, Balkan S, Tamrat A, Priotto G, Alberti KP, Guthmann JP. Malaria epidemics and interventions, Kenya, Burundi, Southern Sudan, and Ethiopia, 1999–2004. Emerg Infect Dis. 2006;10:1477–1485. doi: 10.3201/eid1210.060540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aregawi M, Lynch M, Bekele W, Kebede H, Jima D, Taffese HS, et al. Time series analysis of trends in malaria cases and deaths at hospitals and the effect of antimalarial interventions, 2001–2011, Ethiopia. PLoS One. 2014;9:e106359. doi: 10.1371/journal.pone.0106359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deribew A, Dejene T, Kebede B, Tessema GA, Melaku YA, Misganaw A, et al. Incidence, prevalence and mortality rates of malaria in Ethiopia from 1990 to 2015: analysis of the global burden of diseases 2015. Malar J. 2017;16:271. doi: 10.1186/s12936-017-1919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Federal Ministry of Health Ethiopia (FMOH). Ethiopia National Malaria strategic plan: 2017–2020. Addis Ababa; 2017.
- 7.WHO. World malaria report 2015. Geneva: World Health Organization; 2015. http://apps.who.int/iris/bitstream/10665/200018/1/9789241565158_eng.pdf?ua=1.
- 8.WHO. World malaria report 2017. Geneva: World Health Organization; 2017. http://www.who.int/malaria/publications/world-malaria-report-2017/en/.
- 9.Ochola LB, Vounatsou P, Smith T, Mabaso ML, Newton CR. The reliability of diagnostic techniques in the diagnosis and management of malaria in the absence of a gold standard. Lancet Infect Dis. 2006;6:582–588. doi: 10.1016/S1473-3099(06)70579-5. [DOI] [PubMed] [Google Scholar]
- 10.Ethiopia National Malaria Indicator Survey Report-EMIS 2011. Ethiopian Public Health Institute; 2012. http://www.ephi.gov.et/images/downloads/ethiopia-mis2011-report.pdf.
- 11.Jima D, Getachew A, Bilak H, Steketee RW, Emerson PM, Graves PM, et al. Malaria indicator survey 2007, Ethiopia: coverage and use of major malaria prevention and control interventions. Malar J. 2010;9:58. doi: 10.1186/1475-2875-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ethiopian National Malaria indicator Survey 2015. Ethiopian Public Health Institute; 2016. https://www.ephi.gov.et/images/pictures/download2009/MIS-2015-Final-Report-December-2016.pdf.
- 13.Zaw MT, Thant M, Hlaing TM, Aung NZ, Thu M, Phumchuea K, et al. Asymptomatic and sub-microscopic malaria infection in Kayah State, eastern Myanmar. Malar J. 2017;16:138. doi: 10.1186/s12936-017-1789-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat Rev Microbiol. 2014;12:833–840. doi: 10.1038/nrmicro3364. [DOI] [PubMed] [Google Scholar]
- 15.Rogier E, Wiegand R, Moss D, Priest J, Angov E, Dutta S, et al. Multiple comparisons analysis of serological data from an area of low Plasmodium falciparum transmission. Malar J. 2015;14:436. doi: 10.1186/s12936-015-0955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook J, Reid H, Iavro J, Kuwahata M, Taleo G, Clements A, et al. Using serological measures to monitor changes in malaria transmission in Vanuatu. Malar J. 2010;9:169. doi: 10.1186/1475-2875-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corran P, Coleman P, Riley E, Drakeley C. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol. 2007;23:575–582. doi: 10.1016/j.pt.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 18.Priest JW, Plucinski MM, Huber CS, Rogier E, Mao B, Gregory CJ, et al. Specificity of the IgG antibody response to Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale MSP119 subunit proteins in multiplexed serologic assays. Malar J. 2018;17:417. doi: 10.1186/s12936-018-2566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heppner DG, Kester KE, Ockenhouse CF, Tornieporth N, Ofori O, Lyon JA, et al. Towards an RTS, S-based, multi-stage, multi-antigen vaccine against falciparum malaria: progress at the Walter Reed Army Institute of Research. Vaccine. 2005;23:2243–2250. doi: 10.1016/j.vaccine.2005.01.142. [DOI] [PubMed] [Google Scholar]
- 20.Tongren JE, Drakeley CJ, McDonald SLR, Reyburn HG, Manjurano A, Nkya WMM, et al. Target antigen, age, and duration of antigen exposure independently regulate immunoglobulin G subclass switching in malaria. Infect Immun. 2006;74:257–264. doi: 10.1128/IAI.74.1.257-264.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogier E, Moss DM, Chard AN, Trinies V, Doumbia S, Freeman MC, et al. Evaluation of immunoglobulin G responses to Plasmodium falciparum and Plasmodium vivax in Malian school children using multiplex bead assay. Am J Trop Med Hyg. 2017;96:312–318. doi: 10.4269/ajtmh.16-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SLR, Carneiro I, et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci USA. 2005;102:5108–5113. doi: 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krivoruchko K. Empirical Bayesian Kriging 2012. https://www.esri.com/news/arcuser/1012/empirical-byesian-kriging.html. Accessed 27 Dec 2018.
- 24.Alemu K, Worku A, Berhane Y, Kumie A. Men traveling away from home are more likely to bring malaria into high altitude villages, northwest Ethiopia. PLoS One. 2014;9:e0095341. doi: 10.1371/journal.pone.0095341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yukich JO, Taylor C, Eisele TP, Reithinger R, Nauhassenay H, Berhane Y. Travel history and malaria infection risk in a low-transmission setting in Ethiopia: a case control study. Malar J. 2013;12:33. doi: 10.1186/1475-2875-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asnakew Y, Belendia S, Caterina G, Berhane T, Steffanie C, Teklehaimanot GK, et al. Movement patterns of seasonal migrant workers associated with malaria risk derived from outpatient register books in Amhara Region, Ethiopia. 2017. https://path.org/resources/movement-patterns-of-seasonal-migrant-workers-associated-with-malaria-risk-derived-from-outpatient-register-books-in-amhara-region-ethiopia/.
- 27.Ashton RA, Kefyalew T, Tesfaye G, Pullan RL, Yadeta D, Reithinger R, et al. School-based surveys of malaria in Oromia Regional State, Ethiopia: a rapid survey method for malaria in low transmission settings. Malar J. 2011;10:25. doi: 10.1186/1475-2875-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donahue CG, Carruthers VB, Gilk SD, Ward GE. The Toxoplasma homolog of Plasmodium Apical Membrane Antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol Biochem Parasitol. 2000;111:15–30. doi: 10.1016/S0166-6851(00)00289-9. [DOI] [PubMed] [Google Scholar]
- 29.Scott CA, Yeshiwondim AK, Serda B, Guinovart C, Tesfay BH, Agmas A, et al. Mass testing and treatment for malaria in low transmission areas in Amhara Region, Ethiopia. Malar J. 2016;15:305. doi: 10.1186/s12936-016-1333-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Malaria Control Team, World Health Organization, Ethiopian Addis Ababa University and the INFORM project, Ethiopian Public Health Institute, Department of Public Health Kenya Medical Research Institute. An epidemiological profile of malaria in Ethiopia. 2014.
- 31.White NJ. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011;10:297. doi: 10.1186/1475-2875-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assefa A, Ali A, Deressa W, Tsegaye W, Abebe G, Sime H, et al. Glucose-6-phosphate dehydrogenase (G6PD) deficiency in Ethiopia: absence of common African and Mediterranean allelic variants in a nationwide study. Malar J. 2018;17:388. doi: 10.1186/s12936-018-2538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondrashin A, Baranova AM, Ashley EA, Recht J, White NJ, Sergiev VP. Mass primaquine treatment to eliminate vivax malaria: lessons from the past. Malar J. 2014;13:51. doi: 10.1186/1475-2875-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koepfli C, Colborn KL, Kiniboro B, Lin E, Speed TP, Siba PM, et al. A high force of Plasmodium vivax blood-stage infection drives the rapid acquisition of immunity in Papua New Guinean children. PLoS Negl Trop Dis. 2013;7:e2403. doi: 10.1371/journal.pntd.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woyessa A, Deressa W, Ali A, Lindtjørn B. Prevalence of malaria infection in Butajira area, south-central Ethiopia. Malar J. 2012;11:84. doi: 10.1186/1475-2875-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yalew WG, Pal S, Bansil P, Dabbs R, Tetteh K, Guinovart C, et al. Current and cumulative malaria infections in a setting embarking on elimination: Amhara, Ethiopia. Malar J. 2017;16:242. doi: 10.1186/s12936-017-1884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alemu A, Fuehrer H-P, Getnet G, Tessema B, Noedl H. Plasmodium ovale curtisi and Plasmodium ovale wallikeri in North-West Ethiopia. Malar J. 2013;12:346. doi: 10.1186/1475-2875-12-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong JC. Plasmodium ovale endemic in Ethiopia. Trans R Soc Trop Med Hyg. 1969;63:287–288. doi: 10.1016/0035-9203(69)90164-3. [DOI] [PubMed] [Google Scholar]
- 39.Krafsur ES, Armstrong JC. Epidemiology of Plasmodium malariae infection in Gambela, Ethiopia. Parassitologia. 1982;24:105–120. [PubMed] [Google Scholar]
- 40.WHO. World Malaria Report. Geneva: World Health Organization; 2011. http://www.who.int/malaria/world_malaria_report_2011/en/.
- 41.Woodberry T, Minigo G, Piera KA, Hanley JC, de Silva HD, Salwati E, et al. Antibodies to Plasmodium falciparum and Plasmodium vivax Merozoite Surface Protein 5 in Indonesia: species-specific and cross-reactive responses. J Infect Dis. 2008;198:134–142. doi: 10.1086/588711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birhanu Z, Yihdego YY, Yewhalaw D. Quantifying malaria endemicity in Ethiopia through combined application of classical methods and enzyme-linked immunosorbent assay: an initial step for countries with low transmission initiating elimination programme. Malar J. 2018;17:152. doi: 10.1186/s12936-018-2282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook J, Xu W, Msellem M, Vonk M, Bergstrom B, Gosling R. Mass screening and treatment on the basis of results of a Plasmodium falciparum-specific rapid diagnostic test did not reduce malaria incidence in Zanzibar. J Infect Dis. 2015;211:1476–1483. doi: 10.1093/infdis/jiu655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helb DA, Tetteh KKA, Felgner PL, Skinner J, Hubbard A, Arinaitwe E, et al. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci USA. 2015;112:E4438–E4447. doi: 10.1073/pnas.1501705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Percent malaria seropositivity and Odds Ratio (OR) by sex, age and elevation for each species in malaria endemic areas of Ethiopia, 2014.
Additional file 2. An Interpolated surface map for P. falciparum (left) and P. vivax (right). Empirical bayesian kriging was used to produce the maps from the Annual Parasite Incidence data in Ethiopia (API 2014).
Additional file 3. Regional age-seroconversion plots for antibody responses for Plasmodium falciparum antigens. Y-axis represents probability of being seropositive and the X-axis age. Seroconversion curves represent the rate at which a population become seropositive to specific antigens resulting in seroconversion rates (SCR) or lambda (λ). In each graph points represent age seroprevalence (by deciles), unbroken line represents maximum likelihood curves and broken lines represent the 95% confidence interval. Plots A and B depict the seroconversion curves for P. falciparum antigens response to MSP-1 (A) and AMA-1(B) for the major four regions Tigray, Amhara, Oromia and Southern Nations and Nationalities People’s Regions.
Additional file 4. Regional age-seroconversion plots for antibody responses for P. falciparum antigens. Y-axis represents probability of being seropositive and the X-axis age. Seroconversion curves represent the rate at which a population become seropositive to specific antigens resulting in seroconversion rates (SCR) or lambda (λ). In each graph points represent age seroprevalence (by deciles), unbroken line represents maximum likelihood curves and broken lines represent the 95% confidence interval. Plots A and B depict the seroconversion curves for P. vivax antigens response to MSP-1 (A) and AMA-1 (B) for the major four regions, Tigray, Amhara, Oromia and Southern Nations and Nationalities People’s Region (SNNPR).
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.