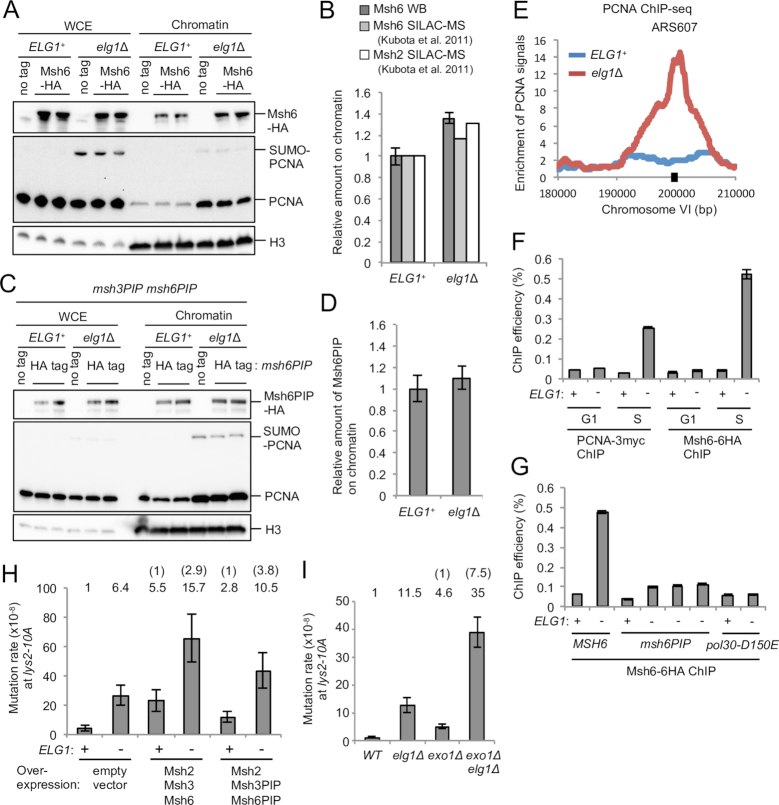
Figure 5.
Over-retained PCNA over-recruits Msh2–Msh6 to chromatin, affecting mismatch repair. (A) Msh6 accumulates on chromatin in elg1Δ, compared to ELG1+. WCE and chromatin-enriched fractions (Chromatin) were prepared from cells expressing Msh6–6HA in log phase. Msh6–6HA, PCNA and histone H3 (loading control) were detected by western blotting. (B) Quantification of chromatin-bound Msh6, normalized to histone H3 (average of two independent clones shown in panel A) and chromatin-bound Msh2 and Msh6 in cells in hydroxyurea by SILAC quantitative proteomics performed previously (46). Error bars, SDs. (C) Msh6PIP does not accumulate on chromatin in elg1Δ, compared to ELG1+. WCE and chromatin-enriched fractions (Chromatin) were prepared from msh6PIP msh3PIP mutants expressing Msh6PIP-6HA in log phase. Msh6PIP-6HA, PCNA and histone H3 (loading control) were detected by Western blotting. (D) Quantification of chromatin-bound Msh6PIP (normalized to histone H3) shown in panel C. Average of two independent clones is shown. Error bars, SDs. (E) ChIP-seq analysis of PCNA performed previously (11). PCNA distribution around ARS607 on chromosome VI is shown. PCNA is unloaded behind replication forks in ELG1+ but retained in elg1Δ in S phase (15 min after release from a cdc7–1 block at 16°C). Black square, region quantified by ChIP-qPCR in panels F and G. (F) ChIP-qPCR analysis of Msh6 and PCNA for early origin ARS607 in the presence and absence of Elg1. ChIP was performed using cells arrested in alpha-factor (G1) or collected 15 min after release from a cdc7–1 block into S phase at 16°C (S). Error bars, SDs of three technical replicates. (G) ChIP-qPCR analysis of Msh6 for ARS607 in the msh6PIP mutant or the disassembly-prone PCNA mutant pol30-D150E backgrounds. ChIP was performed using cells collected 15 min after release from a cdc7–1 block into S phase at 16°C. Three independent isolates of msh6PIP elg1Δ were shown. Error bars, SDs of three technical replicates. (H) Effect of over-expression of Msh2, Msh3 and Msh6 or their PIP mutants on mutation rate at the lys2–10A locus. Msh2, Msh3 and Msh6 or their PIP mutants are over-expressed under their own promoters from three multicopy plasmids. Error bars, 95% confidence intervals. (I) Mutation rates of ELG1+ and elg1Δ in wild-type and exo1Δ backgrounds at the lys2–10A locus. Error bars, 95% confidence intervals.