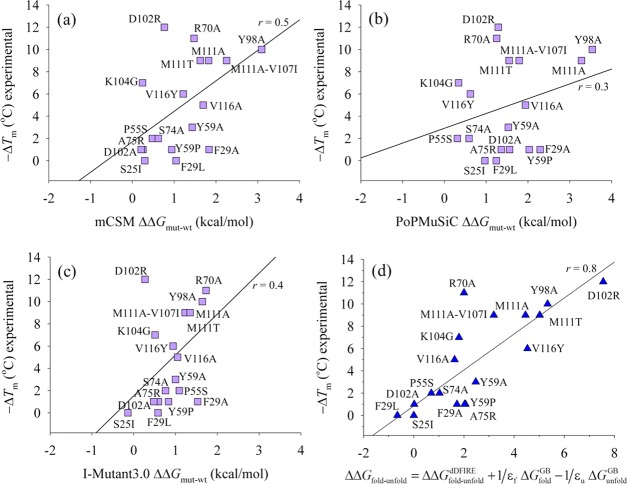
Figure 7.
Modeling 18 sequence substitutions of the B-chain conformer of A3. (a) mCSM server37 predictions showing r ∼ 0.5. (b) PoPMuSiC server18 with r ∼ 0.3. (c) I-Mutant3.0 server16 predictions showing r ∼ 0.4. (d) Linear scaling approximation of scoring mutations by a combined dDFIRE statistical potential and a weighted generalized Born (GB) implicit solvent model applied to a two-state model of the folded and unfolded conformations; the linear regression correlation coefficient is r ∼ 0.8.