Abstract
B-form DNA can adopt alternative structures under conditions such as superhelical duress. Alternative DNA structures are favored when there is asymmetric distribution of guanosine and cytosine on complimentary DNA strands. A guanosine-rich strand can form a four-stranded structure known as a quadruplex (G4). The complimentary cytosine-rich strand can utilize intercalating cytosine–cytosine base pairing to form a four-stranded structure known as the i-motif (iM). Both secondary structures are energetically uphill from double-strand DNA (dsDNA), meaning that additional factors are needed for their formation. Most iMs require slightly acidic conditions for structure stabilization. However, crowding agents such as polyethylene glycols and dextrans can shift the pKa of the iM to near-physiological pH ≈ 7. Nucleic acids have long been known to be bound and stabilized by polyamines such as putrescine, spermidine, and spermine. Polyamines have very high concentrations in cells (0.1–30 mM), and their binding to DNA is driven by electrostatic interactions. Polyamines typically bind in the minor groove of DNA. However, because of the unusual structure of iMs, it was unknown whether polyamines might also bind and stabilize iMs. The study described here was undertaken to analyze polyamine–iM interactions. The thermal stability and pH dependence of iM structures were determined in the presence of polyamines. In contrast to dsDNA, our results suggest that polyamines have considerably weaker interactions with iMs, as demonstrated by the minimal change in iM pH dependence and thermal stability. Our results suggest that polyamines are unlikely to provide a significant source of iM stabilization in vivo.
Introduction
The Watson and Crick B-form DNA structure created the foundation for further investigation into the structure of this iconic biological molecule.1 Subsequent work on DNA’s plasticity resulted in the discovery of various alternative structures, depending on environmental conditions.2 These are usually referred to as secondary structures to differentiate them from B-form DNA. One well-studied secondary structure that is dependent on clustered guanosines and utilization of Hoogsteen base pairing is the quadruple helical structure called a G-quadruplex (G4). Potential G4s have been identified across the genome in telomeres and in promoters of genes often associated with the cell growth, such as bcl-2, ras, VEGF, and c-myc.3−5 G4 structures have significant stability under physiological conditions, allowing for their potential as drug targets. Detection of G4s in cell nuclei has also been reported.6 In biotechnological applications, G4s can also act as reporters for gene deletions7 and for detection of silver, mercury, and other metal ions.8,9
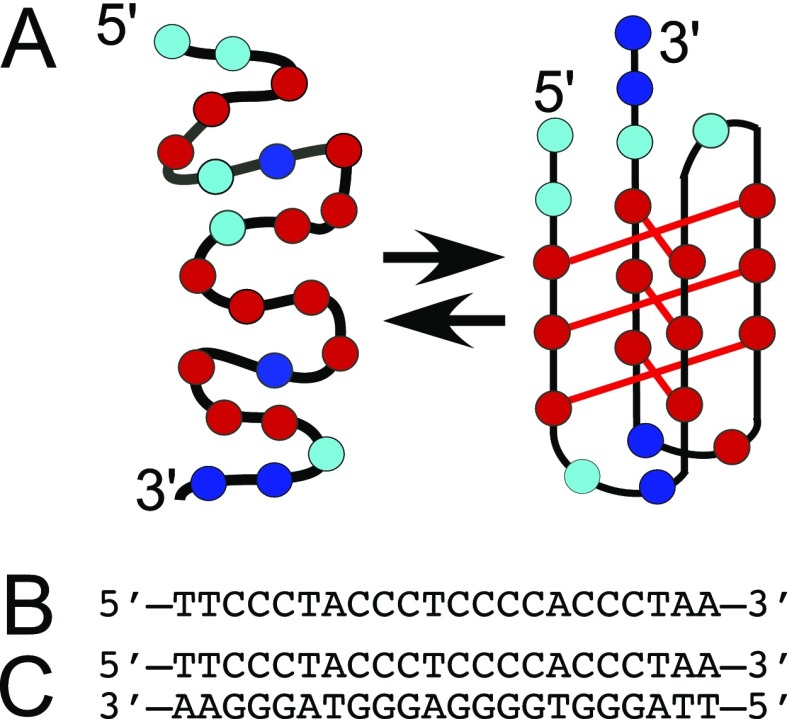
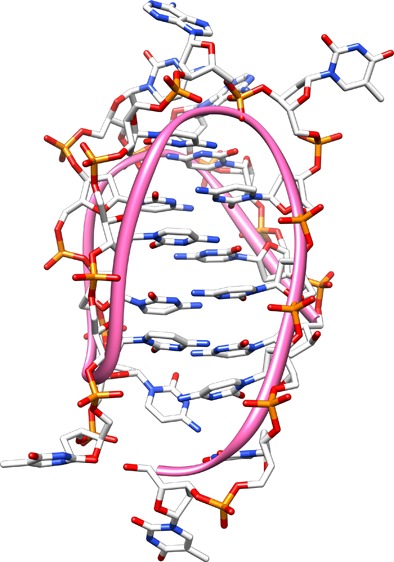
While much is known about the role of G4-forming sequences in biology, much less is known about the complimentary strand that is rich in cytosines. Cytosine-rich single-strand DNA can also form a four-stranded structure. Under slightly acidic conditions (pH ≈ 6.5), the N1 position of cytidine can be protonated, allowing three hydrogen bonds to form between two cytidines.10 The resulting four-stranded structure exhibits intercalated interactions between planes of cytosine base pairs (Figure 2), and therefore, it has been referred to as i-motif (iM) DNA. In early studies and in dilute solutions, at increasing pH, the structural stability of iMs decreases to the point that at physiological pH (∼7.3), little or no iM structure remains.10 Hence, in the past, the iM has attracted less attention than G4s because the nucleus does not appear to be more acidic than the cytoplasm. However, more recent studies have shown that addition of crowding agents and/or dehydrating cosolvents can shift the pKa for the formation of an iM toward more physiological pH.11−14 Longer C-rich sequences that form iMs at pH ≈ 7 have also been reported.15 More recently, iM structures have also been observed in cell nuclei, increasing interest in their possible biological role.16,17
Figure 2.
(A) Folded C6T iM DNA equilibrium. Cyan circles represent dT, red circles dC, and blue circles dA. (B) Single-strand DNA sequence of C6T used for this study. (C) Double-stranded B-form of C6T (dsC6T) used for comparison.
Energetically, it is difficult for G4/iMs to be formed from double-strand DNA (dsDNA). The estimated free energy (ΔG) needed to open the dsDNA to its single strands and form G4/iMs is 20–30 kcal/mol.18 Some of this free energy can be provided by the negative supercoiling of DNA.19,20 Protein binding as the lone stabilizer of the G4/iM structures would require a Kd of ∼10–14 M to provide this ΔG. However, nucleolin binds G4s from the VEGF promoter with Kd ≈ 200 nM, which is energetically too low.21−23 Other proteins that bind preferentially to the G4 structure have also been reported.24,25 Proteins known to bind the iM are the heterogeneous nuclear ribonucleo-protein K and polycytosine binding protein 2, both of which have a high affinity for C-rich strands found in transcriptional promoter regions that are known to form iM structures,26−28 but again the Kd’s are too low to fully support the formation of iMs in vivo. One possible explanation for the observation of iM formation in cells is synergism between supercoiling, protein binding to iMs, and binding of other molecules. This paper addresses the question as to whether polyamines, which are present in high concentrations in cells, might be likely additional enhancers of iM stability.
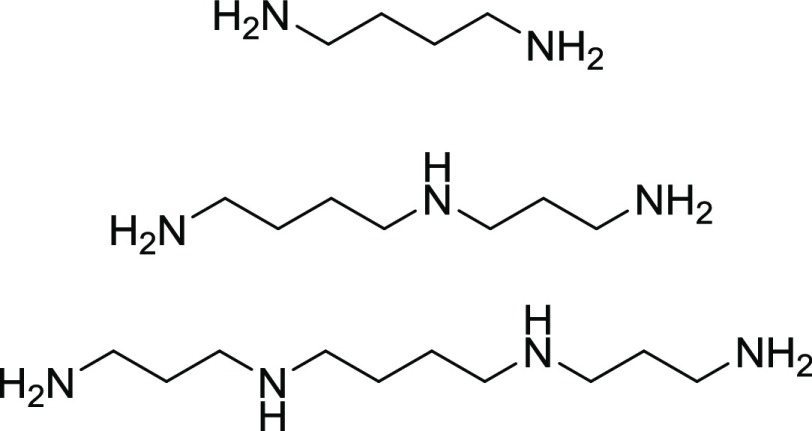
There have been very few studies of the binding of small molecules to iMs. Certain steroids have been reported to selectively bind to regions of DNA that form a hairpin structure in C-rich DNA over the iMs.29 However, a more abundant potential source of iM-stabilizing molecules is polyamines. Polyamines are small biomolecules synthesized in high (0.1–30 mM) concentrations by cells,30 and they are known to be involved in regulation of transcription.31,32 Polyamines bind to the grooves of B-form DNA because of their overall net positive charge at physiological pH. Polyamines such as putrescine, spermidine, and spermine (Figure 1) increase the thermal stability of B-DNA and additionally can induce the transition of B-DNA to left-handed Z-DNA.33
Figure 1.
Molecular structure of the three polyamines used here. From top to bottom: putrescine, spermidine, and spermine. All amino groups are protonated and carry a positive charge at physiological pH (∼7.3) or lower.
To the best of our knowledge, no studies of polyamine binding to iMs have been reported. Hence, in this report, we examined the effect of polyamines on the physical properties of an iM sequence taken from the nuclease-hypersensitive element found within the c-myc promoter (Figure 2). Our study examined the interactions between the c-myc iM and the most prevalent polyamines found in cells (putrescine, spermidine, and spermine). Our results are described below.
Results and Discussion
Polyamine Binding to C6T iM
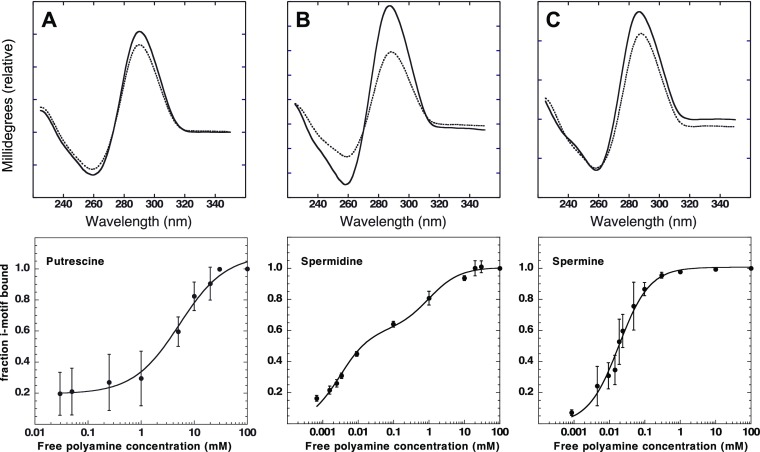
In this study, a fixed concentration of 2 μM of C6T iM was used to determine the binding affinity of each of the three polyamines. Individual samples with a known concentration of polyamine were mixed with a fixed C6T iM concentration. Using electronic circular dichroism (CD) spectroscopy, a titration for each polyamine was observed, as shown in Figure 3. When the concentration of polyamines increased in solution, an increase in the distinct iM CD signal was observed. Saturation was eventually reached. There was distinct variation in the concentrations needed to reach saturation. For putrescine, it was ∼10 mM, while for spermidine and spermine, it was ∼1 and ∼0.05 mM, respectively. This may reflect the length of the polyamine, as our molecular docking (Figure 6) suggests that the grooves of the 6CT iM may accommodate more than one putrescine. There are two nonidentical grooves in the iM (Figure 6). On the basis of the analysis from Figure 3 of the binding plots, the Kd for each polyamine was determined and recorded in Table 1. We note that given the uncertainty of the capacity of the iM to accommodate polyamines, the amount bound is given as a fraction of total iM concentration (Table 2).
Figure 3.
CD spectroscopy of fixed 2 μM C6T iM with increasing polyamine concentrations. Dashed lines are the initial CD signal and solid lines the final CD signal. The panels are for (A) putrescine, (B) spermidine, and (C) spermine. Fits to a single site binding equation for putrescine and spermine are shown. Spermidine was better fit with a two-site model eq 1.
Figure 6.
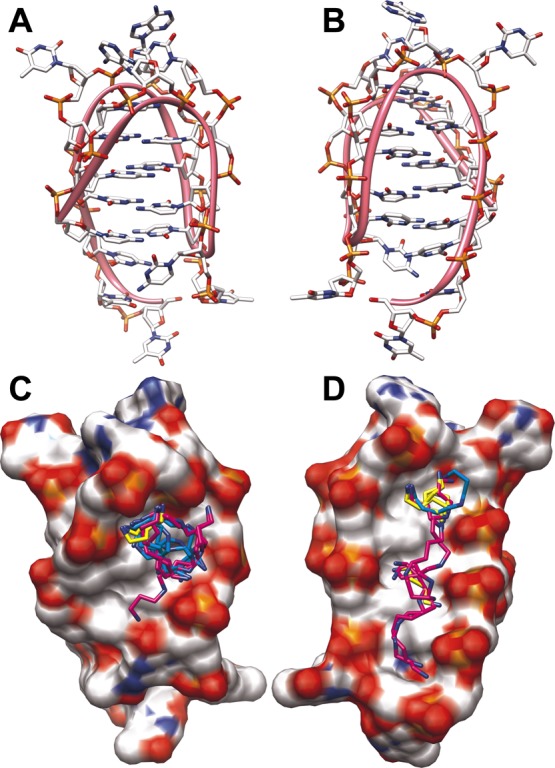
Molecular model of the C6T iM. The structure has two unequal grooves (panels A and B at 90° rotation). The ribbon indicates the backbone trace. The results of molecular docking of polyamines are shown in panels C and D along with space-filling models. Electrostatic charge is shown from positive (blue) to neutral (white) to negative (red). Putrescine is shown in yellow, spermidine in cyan, and spermine in purple.
Table 1. Determined Kd of Polyamine Binding to C6T iM. Parameters Derived from the Data Shown in Figure 3 and Fitting to Eq 1.
| polyamine | n1 | Kd1 (mM) | n2 | Kd2 (mM) |
|---|---|---|---|---|
| putrescine | 0.89 ± 0.04 | 5.3 ± 1.1 | ||
| spermidine | 0.60 ± 0.04 | 0.003 ± 0.001 | 0.40 ± 0.04 | 1.0 ± 0.92 |
| spermine | 1.00 ± 0.02 | 0.018 ± 0.003 |
Table 2. Quantification of Protons Required for C6T Formation without and with Saturating Polyamines Presenta.
| average protons | |
|---|---|
| no polyamines | 2.8 ± 0.9 |
| 10 mM putrescine | 3.2 ± 0.5 |
| 1 mM spermidine | 4.2 ± 0.4 |
| 0.05 mM spermine | 5.6 ± 0.5 |
iM Dependence on Proton Concentration
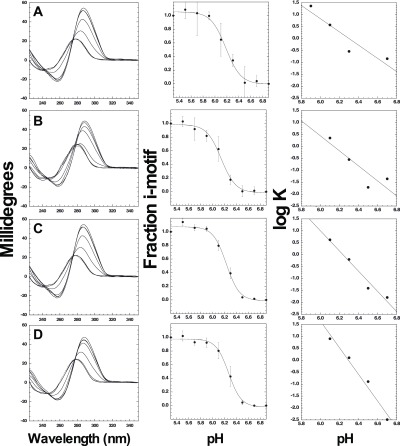
As shown in Figure 4, we used CD spectroscopy to determine the pKa for folding of the C6T iM in the presence of saturating concentrations of polyamines. Interestingly, there was no shift in the pKa observed with any polyamine; the pKa for iM folding under all conditions was pH 6.2 ± 0.1. We analyzed the transition region of the pH-dependent plots using a Wyman plot (Figure 4), which allowed determination of the proton uptake required to transition from unfolded DNA to the iM in the presence or absence of polyamines. In the absence of polyamines, the proton requirement to fold the iM is approximately 3 protons per iM. This is consistent with prior literature where slightly less than half of the cytosines in an iM need to be protonated to stabilize the structure.34 Putrescine at high concentrations has little or no effect on the proton requirement. However, an increase in protons required for stabilization of the C6T iM is observed in the presence of saturating amounts of spermine (∼4) and spermidine (∼6). Our interpretation of these data is that as both spermine and spermidine have long charged chains, their binding interferes with protonation of the cytidines, likely due to charge neutralization of the phosphate backbone. The net result is a requirement of additional protons to stabilize the iM. This manifests in a more cooperative transition from the unfolded to folded state, but the midpoint pH of the transition remains constant.
Figure 4.
pH dependence of iM folding. Row (A) CD spectra for C6T when no polyamines are present at varying pH. In saturating amounts of polyamines: row (B) in 10 mM putrescine; row (C) in 1 mM spermidine; and row (D) in 0.05 mM spermine. Final fractions other than 1.0 are due to fitting, where the final fraction was allowed to be ∼1.0.
Thermal Stability of C6T iM in the Presence of Polyamines Using UV–Vis Spectroscopy
The thermal stability of the C6T iM was observed using UV–vis spectroscopy. The baseline C6T iM thermal melting temperature (TM), which is defined as the temperature when half of its concentration is in the folded state (Figure 5), was measured at pH 5.3 where the C6T iM is most stable. The observed TM in the absence of polyamines was 45.4 ± 0.2 °C (Table 3).
Figure 5.
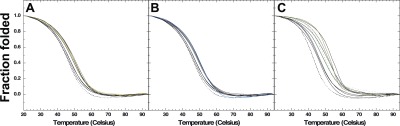
Thermal denaturation of the C6T iM in the presence of varying concentrations of polyamines: (A) putrescine, (B) spermidine, and (C) spermine at pH 5.3.
Table 3. C6T iM (C6T iM) and Double-Strand C6T (dsC6T) Melting Temperature (TM) at pH 5.3.
| TM (°C) | ||||
|---|---|---|---|---|
| DNA | no polyamine | 10 mM putrescine | 1.0 mM spermidine | 0.050 mM spermine |
| C6T iM | 45.4 ± 0.2 | 49.3 ± 0.1 | 53.0 ± 0.1 | 53.1 ± 0.8 |
| dsC6T | 50.4 ± 0.4 | 73.6 ± 0.1 | 76.7 ± 0.4 | 72.8 ± 0.6 |
At saturating concentrations of each of the polyamines and 2 μM C6T, the TM was determined and recorded in Table 3. A slight increase in TM was observed for all of the polyamines: ∼4 °C for putrescine and ∼8 °C for spermidine and spermine. For comparison, we also examined the effects on dsC6T. Consistent with prior literature,35 all polyamines have a pronounced increase (23–27 °C) on the TM of dsDNA. We interpret the difference between iM and dsDNA as a function of the size and shape of the grooves in the two structures. While dsDNA has a long, narrow minor groove, iMs have two wide grooves (Figure 6). Binding of polyamines to dsDNA leads to significant charge neutralization of the phosphate backbone and thus enhances thermal stability. In contrast, polyamine binding to iM DNA appears to be mainly located in the loops of the iM (Figure 6) and hence has little effect on the stability of the core four-stranded structure.
Molecular Modeling of Polyamine Binding to iM DNA
We used AutoDock Vina to explore the possible ways in which polyamines might bind to the C6T iM. Our results are shown in Figure 6 and provide a good model to explain the binding data from Figure 3. Because of the intercalated cytosines, the two grooves in an iM are very wide compared to B-form dsDNA. The most likely location for putrescine is very near the loops of the iM (Figure 6C,D). This likely explains the lower affinity for putrescine for the iMs, inasmuch as the binding locations are limited because it is the shortest polyamine used. If the docking model is correct, there are essentially only two binding sites that favor putrescine. In contrast, the longer spermidine can occupy both the loop regions occupied by putrescine, as well as extending into the groove of the iM (Figure 6C). This would explain the biphasic binding curve of spermidine, where binding to the loop not only has a fractional component with affinity similar to putrescine but also has a groove binding component similar to spermine (see Table 1). Finally, our docking results indicate that spermine, the longest polyamine, has the appropriate molecular dimensions to fit along both grooves of the iM (Figure 6C,D). This would explain the much higher binding affinity of spermine to the iM (Figure 3; Table 1).
Conclusions
In this paper, we concluded that although polyamines bind to iM structures, their binding is weak compared to the more favorable binding of polyamines to the same sequence in the double-strand form of DNA. Using the nuclease-hypersensitive element found within the c-myc gene, denoted as C6T, a distinct iM structure was formed without the presence of polyamines, which had a melting temperature of 45.4 ± 0.2 °C at acidic pH, well above the physiological temperature of 37 °C. Using CD spectroscopy, a titration curve at pH 5.3 of varying concentrations of polyamines was produced, allowing concentration saturations levels to be determined. These results indicated that putrescine, spermidine, and spermine could reach binding saturation to the C6T iM at concentrations of 10, 1, and 0.05 mM, respectively. Using the data collected from CD, the Kd of polyamines’ binding to the C6T iM were determined. The polyamines produced small but significant shifts in the thermal stability of the iM, increasing its TM by 4–8 °C, which is in stark contrast to their ability to shift dsDNA by increasing its TM by ∼23 °C.
It was determined that at saturating concentrations of polyamines, the change in the required amount of protons to stabilize the C6T iM structure is increased. Putrescine displays a small increase in required protons possibly because of the small nature of putrescine as a biomolecule. However, both spermidine and spermine required additional protons, indicating that they may affect the ability of the core cytidines of the iM to form the four-stranded structure. However, no effect on the pKa required to form the iM was observed. Hence, the effect of polyamines appears to be solely on the cooperativity of folding.
Overall, it was determined that the most abundant cellular polyamines putrescine, spermidine, and spermine have the ability to bind to the C6T iM structure, albeit weakly, and to add some stability to the structure. However, none of our data suggests that polyamines have a particularly important role in stabilizing iM structures in vivo. Because of their high concentrations in cells, the polyamines may contribute a certain level of stability to iMs but nowhere near the 20–30 kcal/mol required to stabilize an iM/G4 from opened dsDNA. Hence, other factors that stabilize iMs are part of the ongoing investigation in understanding their physical properties in cells. While our results pertain solely to the cytosine-rich strand of DNA, we anticipate that our findings are readily transferable to the situation where iMs are formed in dsDNA.
Materials and Methods
Materials
The iM-forming oligonucleotide (C6T; Figure 2) used was synthesized by and purchased from Midland Certified Reagent Co., Inc. (Midland, TX). C6T is a good model iM because it undergoes a simple two-state transition from folded to unfolded form.36 C6T iM oligonucleotide stock was stored in a 10 mM Tris, 1 mM ethylenediaminetetraacetic acid (EDTA) buffer at pH 8.0 in a −20 °C freezer. The sodium cacodylate, Tris-HCl, and EDTA used to create buffer solutions were purchased from Fisher Scientific (Pittsburgh, PA). Putrescine, spermidine, and spermine were purchased from Sigma Aldrich (St. Louis, MO).
Polyamine Binding Constants
Binding of polyamines was monitored by electronic CD spectra of the iM DNA. Addition of polyamines resulted in a strong increase in CD signal at 290 nm, which appeared to show typical saturation at higher concentrations. We analyzed this signal change via transformation of the data into a binding curve analysis (Figure 3). The binding plots for putrescine and spermine showed a standard sigmoidal curvature, which could be well fitted to a single site binding model. However, the binding of spermidine was better fit with a model of two classes of independent binding sites (n1, n2) with independent binding constants Kd1 and Kd2 (e.g., the Klotz model).37 The spermidine data were fit to eq 1, where r is the fraction of iM with the ligand bound and L is the free polyamine concentration
| 1 |
pKa Determination
Samples were prepared in the same way as for binding analysis. Both polyamine binding and pKa (the midpoint of the pH-dependent folding for iM) were determined using CD spectroscopy. To determine the pKa for iM folding (i.e., the pH at which 50% of the oligo is folded into the iM), CD spectra of DNA solutions at 20 °C were collected from 250–320 nm on an Olis DSM 20 CD instrument fitted with a Peltier heat block (Olis, Inc. Bogart, GA, USA). An integration time as a function of high voltage was used. The CD signals observed at 298 nm were then plotted against pH, and then eq 2 was applied to obtain pKa and the cooperativity parameter. The cooperativity parameter reflects the slope of the transition from folded to unfolded iM. A technical caution: as polyamines are weak bases and are used at high concentrations, we emphatically note that the pH of buffers containing polyamines have to be adjusted to the correct pH for each concentration of polyamine used.
| 2 |
To better understand the cooperativity parameter, a Wyman-type plot34,38 was used to determine the proton difference (ΔQ) between the folded and unfolded iM. ΔQ can be determined via the slope of each of the Wyman plots of log K versus pH, where K is the equilibrium constant between unfolded and folded iM (defined in eq 5 below)
| 3 |
Thermal Denaturation Studies
Relative to B-form or single-strand DNA, the iM structures show a pronounced hypochromic and bathochromic shift in their UV–visible absorbance spectra that can be monitored at 260 nm to determine folded and unfolded states (Figure S1). Solutions of the C6T iM oligonucleotide (2 μM) in 30 mM cacodylate buffer at pH 5.3 were prepared with varying concentrations of polyamines. Melting of the C6T iM with and without polyamines was done using a Cary 100 UV–visible spectrometer (Agilent Technologies, Santa Clara, CA). Prior to experimentation, each sample was heated to 80 °C for 5 min and then cooled to room temperature for two cycles to ensure removal of mismatched DNA dimers. Thermal denaturation recordings were made by monitoring the absorbance at 260 nm while increasing the temperature between 20 and 94 °C at a ramping rate of 2 °C per min and a 1 min hold at each temperature. All thermal denaturation experiments could be fit to a simple two-state model.
The two-state model for DNA melting is described by
| 4 |
and
| 5 |
where K is the equilibrium constant for unfolding, [U] and [N] are the concentrations of unfolded and folded states, respectively, and T is the temperature in kelvin at each point along the melting transition.
The mole fraction of unfolded DNA f(U) is given by
| 6 |
The free energy of unfolding at any given temperature T is given by the following
| 7 |
The fraction folded was normalized from 0 to 1 prior to fitting, and the change in heat capacity (ΔCp) was assumed to be negligible. The nonlinear regression fits yielded the values for Tm and ΔHm. Note that in eq 7, ΔHm reflects the slope of the thermal transitions shown in Figure 5. Because there were no significant differences in slopes for all the conditions used, we have not reported separate ΔHm values, but its average value was ∼33 kcal/mol.
Visualization of the Best-Predicted Polyamine Binding Location
Using the open source program AutoDock Vina,39 the most likely locations of molecular docking of the polyamines to the C6T iM were determined. The model of C6T used was built by us and has been described previously.40 The polyamine torsion angles were set to move freely to simulate their flexibility. Both the iM and polyamine structures were assigned charges found at physiological pH. The output was visualized using UCSF CHIMERA.41
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b00784.
Change in absorbance for fully folded C6T iM DNA at 20 °C and fully unfolded DNA at 80 °C (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Watson J. D.; Crick F. H. C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Choi J.; Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley L. H.; Wheelhouse R. T.; Sun D.; Kerwin S. M.; Salazar M.; Fedoroff O. Y.; Han F. X.; Han H.; Izbicka E.; Von Hoff D. D. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000, 85, 141–158. 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- Brooks T. A.; Hurley L. H. Targeting MYC Expression through G-Quadruplexes. Genes Cancer 2010, 1, 641–649. 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G.; Tannahill D.; McCafferty J.; Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; He B.; Lu L.; Leung C.-H.; Mergny J.-L.; Ma D.-L. Label-free luminescent detection of LMP1 gene deletion using an intermolecular G-quadruplex-based switch-on probe. Biosens. Bioelectron. 2015, 70, 338–344. 10.1016/j.bios.2015.03.047. [DOI] [PubMed] [Google Scholar]
- Ma D.-L.; Lin S.; Lu L.; Wang M.; Hu C.; Liu L.-J.; Ren K.; Leung C.-H. G-quadruplex-based logic gates for HgII and AgI ions employing a luminescent iridium(iii) complex and extension of metal-mediated base pairs by polymerase. J. Mater. Chem. B 2015, 3, 4780–4785. 10.1039/c5tb00718f. [DOI] [PubMed] [Google Scholar]
- Leung K.-H.; He B.; Yang C.; Leung C.-H.; Wang H.-M. D.; Ma D.-L. Development of an Aptamer-Based Sensing Platform for Metal Ions, Proteins, and Small Molecules through Terminal Deoxynucleotidyl Transferase Induced G-Quadruplex Formation. ACS Appl. Mater. Interfaces 2015, 7, 24046–24052. 10.1021/acsami.5b08314. [DOI] [PubMed] [Google Scholar]
- Gehring K.; Leroy J.-L.; Guéron M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Bhavsar-Jog Y. P.; Van Dornshuld E.; Brooks T. A.; Tschumper G. S.; Wadkins R. M. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry 2014, 53, 1586–1594. 10.1021/bi401523b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J.; Waltman P.; Le V.; Lewis E. The effect of molecular crowding on the stability of human c-MYC promoter sequence i-motif at neutral pH. Molecules 2013, 18, 12751–12767. 10.3390/molecules181012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran A.; Nakano S.-i.; Sugimoto N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010, 46, 1299–1301. 10.1039/b922050j. [DOI] [PubMed] [Google Scholar]
- Reilly S. M.; Morgan R. K.; Brooks T. A.; Wadkins R. M. Effect of Interior Loop Length on the Thermal Stability and pKa of i-Motif DNA. Biochemistry 2015, 54, 1364–1370. 10.1021/bi5014722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. P.; Huppert J. L.; Waller Z. A. E. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017, 45, 2951–2959. 10.1093/nar/gkx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeraati M.; Langley D. B.; Schofield P.; Moye A. L.; Rouet R.; Hughes W. E.; Bryan T. M.; Dinger M. E.; Christ D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- Dzatko S.; Krafcikova M.; Hänsel-Hertsch R.; Fessl T.; Fiala R.; Loja T.; Krafcik D.; Mergny J.-L.; Foldynova-Trantirkova S.; Trantirek L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew. Chem., Int. Ed. 2018, 57, 2165–2169. 10.1002/anie.201712284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Hurley L. H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C.; Cui Y.; Mao H.; Hurley L. H. A Mechanosensor Mechanism Controls the G-Quadruplex/i-Motif Molecular Switch in the MYC Promoter NHE III1. J. Am. Chem. Soc. 2016, 138, 14138–14151. 10.1021/jacs.6b09196. [DOI] [PubMed] [Google Scholar]
- González V.; Guo K.; Hurley L.; Sun D. Identification and Characterization of Nucleolin as a c-mycG-quadruplex-binding Protein. J. Biol. Chem. 2009, 284, 23622–23635. 10.1074/jbc.m109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Hurley L. H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe D. J.; Guo K.; Shin Y.-J.; Sun D. Heterogeneous Nuclear Ribonucleoprotein K and Nucleolin as Transcriptional Activators of the Vascular Endothelial Growth Factor Promoter through Interaction with Secondary DNA Structures. Biochemistry 2011, 50, 3796–3806. 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B.; Mayer C.; Varshney D.; Marsico G.; Murat P.; Taylor C.; D’Santos C.; Tannahill D.; Balasubramanian S. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018, 46, 11592–11604. 10.1093/nar/gky861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M.; Singh M. RGG-box in hnRNPA1 specifically recognizes the telomere G-quadruplex DNA and enhances the G-quadruplex unfolding ability of UP1 domain. Nucleic Acids Res. 2018, 46, 10246–10261. 10.1093/nar/gky854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T.; Levens D. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 5830–5835. 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix L.; Lienard H.; Labourier E.; Djavaheri-Mergny M.; Lacoste J.; Leffers H.; Tazi J.; Helene C.; Mergny J. L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000, 28, 1564–1575. 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.-J.; Kendrick S.; Hecht S. M.; Hurley L. H. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014, 136, 4172–4185. 10.1021/ja4109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick S.; Kang H.-J.; Alam M. P.; Madathil M. M.; Agrawal P.; Gokhale V.; Yang D.; Hecht S. M.; Hurley L. H. The Dynamic Character of the BCL2 Promoter i-Motif Provides a Mechanism for Modulation of Gene Expression by Compounds That Bind Selectively to the Alternative DNA Hairpin Structure. J. Am. Chem. Soc. 2014, 136, 4161–4171. 10.1021/ja410934b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.; Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- Thomas T.; Thomas T. J. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 2001, 58, 244–258. 10.1007/pl00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs A. C.; Mehta D. J.; Gerner E. W. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 2003, 60, 1394–1406. 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu H. S.; Marton L. J. The interaction of spermine and pentamines with DNA. Biochem. J. 1987, 244, 243–246. 10.1042/bj2440243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.; Fraire C.; Sheardy R. D. Linking pH, Temperature, and K+ Concentration for DNA i-Motif Formation. J. Phys. Chem. B 2017, 121, 7872–7877. 10.1021/acs.jpcb.7b06317. [DOI] [PubMed] [Google Scholar]
- Bloomfield V. A.; Crothers D. M.; Tinoco I. Jr.. Nucleic acids: structures, properties, and functions; University Science Books: Sausalito, California, 2000. [Google Scholar]
- Dai J.; Hatzakis E.; Hurley L. H.; Yang D. I-Motif Structures Formed in the Human c-MYC Promoter Are Highly Dynamic-Insights into Sequence Redundancy and I-Motif Stability. PLoS One 2010, 5, e11647 10.1371/journal.pone.0011647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz I. M.Ligand-Receptor Energetics; Wiley-Interscience: New York, 1997. [Google Scholar]
- Di Cera E.; Gill S. J.; Wyman J. Canonical formulation of linkage thermodynamics. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 5077–5081. 10.1073/pnas.85.14.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly S. M.; Lyons D. F.; Wingate S. E.; Wright R. T.; Correia J. J.; Jameson D. M.; Wadkins R. M. Folding and Hydrodynamics of a DNA i-Motif from the c-MYC Promoter Determined by Fluorescent Cytidine Analogs. Biophys. J. 2014, 107, 1703–1711. 10.1016/j.bpj.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.