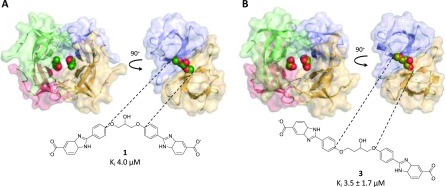
Figure 3.
Crystal structures of DfrB1 (A) with inhibitor 1 PDB ID 6NY0 and (B) with inhibitor 3 PDB ID 6NXZ. Because of the ambiguity of the electron density in the asymmetric unit, we were unable to definitely assign one enantiomer of the racemic central core of 3. We modeled the R-enantiomer in the active site, for further analysis. As the binding modes of 1 and 3 are slightly offset from the center of symmetry, they are observed in two slightly different binding modes when the asymmetric unit is expanded to show the tetramer. The inhibitors were modeled at 0.5 occupancy per asymmetric unit. This approximated two fully occupied binding sites in the tetrameric active-site tunnel.