Figure 3.
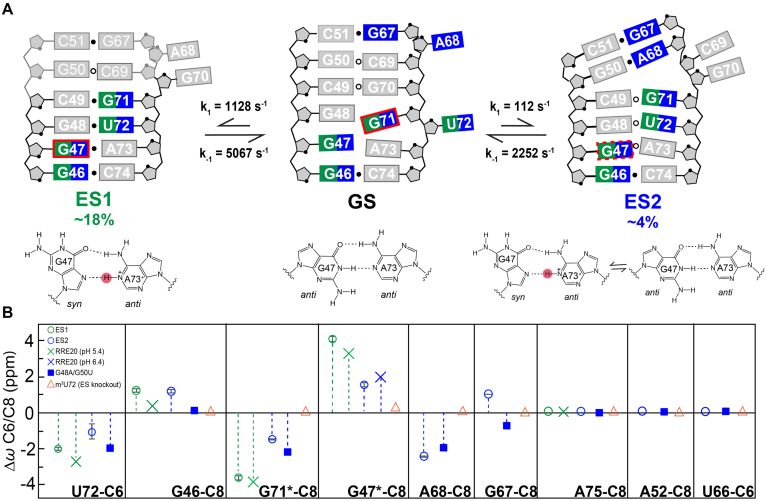
Chemical shift fingerprinting RRE ESs. (A) Putative secondary structure of RREIIB ES1 and ES2. Nucleotides that experience exchange due to ES1 and ES2 are colored green and blue, respectively. Syn bases are indicated with red boxes. The G47(syn)–A73+(anti) ⇌ G47(anti)–A73(anti) equilibrium indicated with the red-dashed box on ES2. The different G47–A73 conformations in GS, ES1 and ES2 are shown below the secondary structures. Base pairs with and without detectable H-bonds are indicated using filled and empty circles, respectively. (B) Comparison of Δω = ωES – ωGS obtained using RD (values obtained in the absence of Mg2+ indicated using asterisk) with corresponding Δωmut = ωES-mutant – ωwild-type obtained for the mutants. Buffer conditions: 15 mM sodium phosphate, 25 mM NaCl, 0.1 mM EDTA, pH 6.4 with or without 3 mM MgCl2.