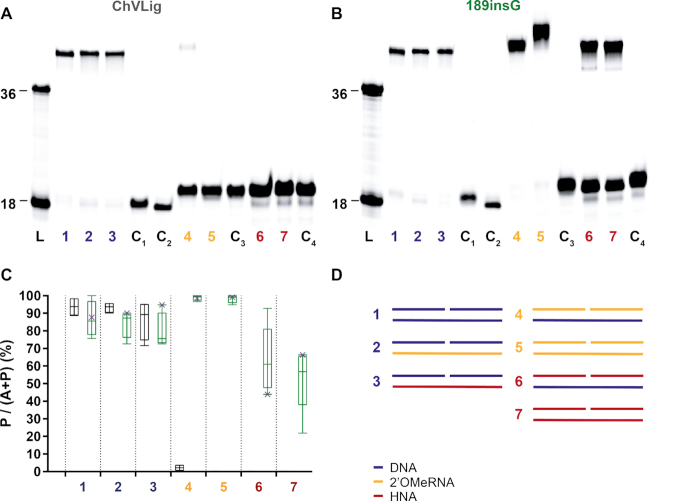
Figure 4.
Ligation of DNA, 2′OMeRNA and HNA fragments by ChVLig (panel A) and its 189insG mutant (panel B) on different complementary templates. Panels a and b display typical PAGE gels for ligation efficiency. On both PAGE, lane L displays a ladder marked at 18 and 36 oligonucleotide length (Supplementary information S7.1, substrates 3a and 7b), lanes indicated with C are negative controls for template independent ligation (lane C1), enzyme independent DNA ligation in the presence of a DNA template (lane C2), enzyme independent 2′OMeRNA ligation in the presence of a 2′OMeRNA template (lane C3) and enzyme independent HNA ligation in the presence of a HNA template (lane C4). The ligation reactions are shown in panel D, where DNA, 2′OMe-RNA and HNA segments are colored in blue, gold and red respectively. The diagram (panel C) summarizes results quantitatively (ChVLig – gray and 189insG –green). Minimum and maximum ligation efficiency are shown as interval bars (n = 4). Representative points of ligation efficiencies from panels a and b are marked with x. Almost full 2′OMeRNA ligation on both DNA and 2′OMeRNA templates occurs compared to a virtually non-existing activity for ChVLig. HNA ligation efficiency reaches on average 63 (±19)% and 53 (±18)% on DNA and HNA templates respectively, compared to non-measurable HNA ligation by the wild-type enzyme.