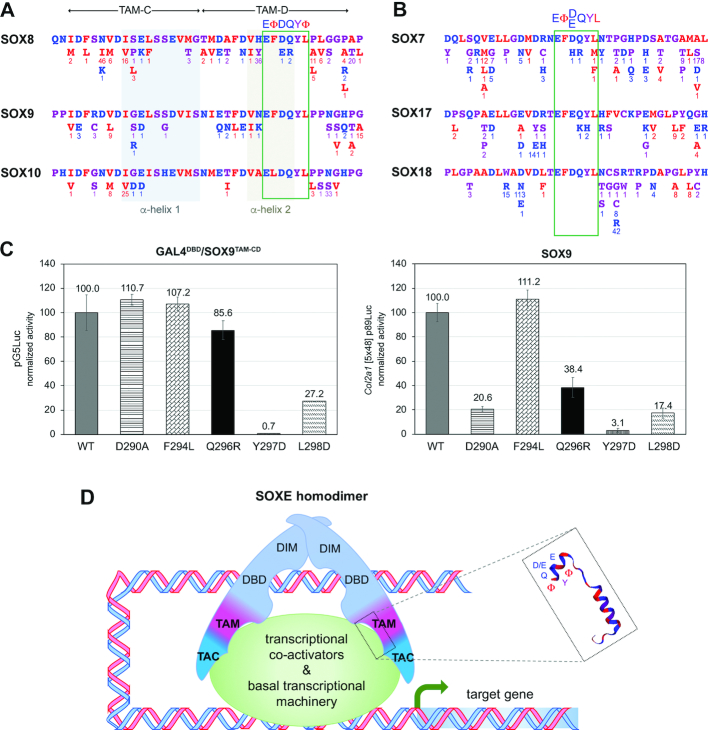
Figure 8.
Analysis of SOXE and SOXF missense variants in the human control population and model for the SOXE bipartite transactivation mechanism. (A) Missense variants listed in gnomAD in SOXE TAM-CD are presented underneath the domain sequences. The numbers of alleles detected in over 140 000 unrelated individuals are indicated for each variant. (B) Missense variants listed in gnomAD in the SOXF EΦ[D/E]QYL motif and flanking residues. (C) Test of the effect of a Q296R variant detected in SOX8 in healthy human individuals on transactivation. The variant was introduced in the GALDBD/SOX9TAM-CD and SOX9 proteins. The proteins were then tested in HEK-293 cells upon co-transfection with the pG5Luc or Col2a1 reporter. Other missense variants (as described in Figure 6) were tested in parallel for comparison. Data were calculated and are presented as in similar assays in previous figures. They were reproduced in multiple experiments. (D) Model of the current view for the mechanism used by SOXE proteins to activate target genes. Previous studies demonstrated that SOXE proteins homodimerize upon binding to inverted recognition sites present in the enhancers or promoter of their target genes. These events involve their HMG and dimerization domains. The present study uncovered that they transactivate these genes through a bi-partite mechanism involving a centrally located domain (TAM) synergizing with the protein C-terminal region (TAC). The functional core of TAM is predicted to fold into binding pocket-like structure upon interaction with specific transcriptional co-activators or components of the basal transcription machinery. It contains a highly conserved EΦ[D/Q]YΦ motif that is critical for transactivation and is thus likely involved in recognizing and firmly binding functional partners. Not shown here is a PQA-rich domain present solely in SOX9 that may facilitate transactivation in specific contexts.