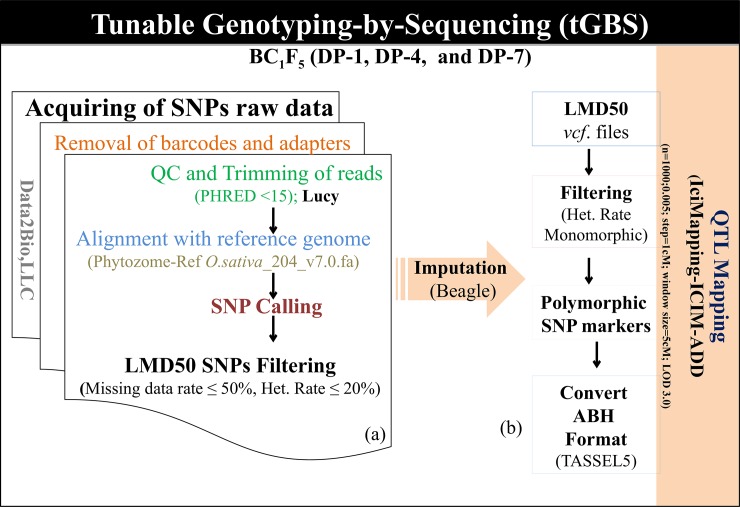
Fig 1. A diagramatic representation of the work-flow of tGBS data analysis.
(a) Trimming of nucleotide raw sequence reads, SNP discovery, SNP calling, and removal of low-quality SNPs were performed by Ali et al. [73]. (b) The LMD50 SNP files (low missing SNP data rate of ≤50%) of three genetic backgrounds (DP-1, DP-4, and DP-7 MPs) were filtered SNPs and converted to ABH format for QTL mapping studies.