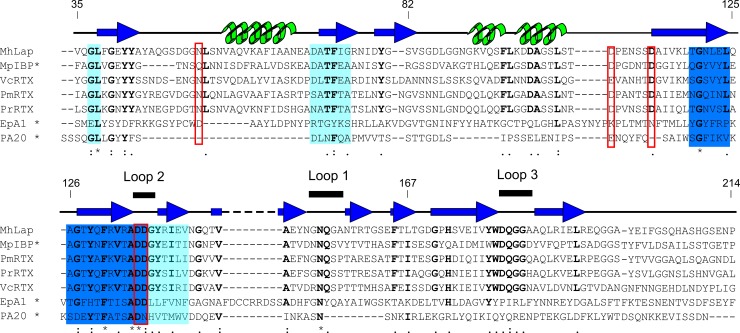
Fig 2. Sequence alignment of PA14 domains.
Sequences of PA14 domains from Marinobacter hydrocarbonoclasticus (MhLap), Marinomonas primoryensis (MpIBP), Pseudomonas mendocina and resinovorans (PmRTX and PrRTX, respectively), Vibrio cholerae (VcRTX), Candida glabrata (EpA1), and Bacillus anthracis (PA20) were aligned using the MergeAlign software [45]. Sequences with alignment scores above 60% are coloured dark blue, while sequences with alignment scores between 30 and 40% are shown in light blue. Proposed Ca2+-binding residues are boxed with red squares. Residue conservation is indicated as follows: * = 100%,: = ~80%,. = ~70%. The conserved residues are bolded. Residue numbers for the MhPA14 construct are given. Secondary structure taken from the solved structure of MpIBP PA14 domain is shown above the alignment, with beta-strands coloured dark blue, alpha-helices coloured green, and loops coloured black. Bold lines sit above the sequence for three loops that make up the supposed sugar-binding site of the bacterial PA14 domains. Note: sequences with previously solved protein structures are marked with an asterisk.