Abstract
Background
Circular RNAs (circRNAs) play important roles in the progression of cancers, but the precise role of circRNAs in the diagnosis and prognosis of hepatocellular carcinoma (HCC) remains to be clarified. The aim of the current study was to explore the diagnostic and prognostic values of hsa_circ_0003998 in HCC.
Methods
CircRNAs expression was measured using RNA-seq analysis from HCC tissues (n=6) (three cases with or without portal vein invasion). Hsa_circ_0003998 in 200 pairs of HCC and adjacent noncancerous tissues and HCC cell lines was examined using qRT-PCR and the clinicopathologic significance was determined. We also detected the plasma levels of hsa_circ_0003998 in HCC, hepatitis B patients and healthy controls. The clinical diagnosis and prognostic values were further determined using receiver operating characteristic (ROC) curve, Kaplan–Meier curve and Cox regression.
Results
Hsa_circ_0003998 was upregulated in HCC tissues (P<0.001) and HCC cell lines (HepG2, HuH7, MHCC97H) (P<0.001). In addition, upregulation of hsa_circ_0003998 level was associated with higher serum alpha-fetoprotien (AFP) level (P=0.003), larger tumor diameter (P=0.009), lower differentiation level (P=0.023) and microvascular invasion (P=0.028). The plasma level of hsa_circ_0003998 in HCC patients was significantly higher than those in hepatitis B patients (P<0.001) and healthy controls (P<0.001). Its level was significantly reduced after the operation (P<0.001). The area under the ROC curve (AUC) for distinguishing HCC from adjacent noncancerous tissues was 0.894 (95% CI=0.86–0.922, P<0.001), the sensitivity and specificity were 0.84 and 0.8, respectively. Comparing with hepatitis B patients and healthy controls, hsa_circ_0003998, respectively, had an AUC value of 0.833 (95% CI=0.763–0.889, P<0.001) and 0.892 (95% CI=0.831–0.937, P<0.001). Their sensitivity and specificity were 0.83, 0.7 and 0.8, 0.84, respectively. Moreover, the combination of hsa_circ_0003998 and AFP showed the highest AUC value of 0.947, the sensitivity and specificity were 0.88 and 0.92, respectively. The hsa_circ_0003998 (P=0.003) and AFP (P=0.008) levels were independent prognostic factors for HCC. The overall survival of HCC patients with high level of hsa_circ_0003998 was significantly poorer than those with low level (P=0.005).
Conclusion
Our findings suggest that hsa_circ_0003998 may be used as a novel potential biomarker for the diagnosis and prognosis of HCC patients.
Keywords: hepatocellular carcinoma, circular RNA, hsa_circ_0003998, biomarker, diagnosis, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the most common malignancy of the liver and the fourth leading cause of cancer-related mortality wordwide.1,2 Although surgical resection is still the primary approach for HCC patients, only 30–40% patients are operable, partially due to the factor that majority of HCC patients were diagnosed at the advanced stage.3 Alpha-fetoprotien (AFP) is the most commonly used biomarker for diagnosis of HCC. However, the sensitivity and specificity of AFP are unsatisfied for screening of early-stage HCC.4 Thus, it is imperative to identify novel biomarkers for HCC diagnosis.
Circular RNA (circRNA) is a type of endogenous noncoding RNA with a stable structure formed by special loop splicing.5 It was regarded as non-functional by-products of splicing errors.6 Recently, mainly because of improvement in high-throughput sequencing and bioinformatics, a growing number of circRNAs has been discovered in a number of cancers.7,8 CircRNAs may act as miRNA sponges, protein-binding molecules and templates for translation into polypeptides in the regulation of physiopathology.9–11 Increasing studies demonstrate that circRNA-mediated networks involved in cell apoptosis, proliferation, cell cycle control and metastasis in cancer.12–15 Furthermore, circRNAs might also act as potential novel diagnostic and prognostic biomarkers of cancer.16,17
In the present study, we utilized RNA-sequencing to determine if there were differentially expressed circRNAs in six cases of HCC tissues (three cases with or without portal vein invasion). We found that hsa_circ_0003998 expression in HCC tissues with portal vein invasion was significantly higher than those without portal vein invasion. Hsa_circ_0003998 is a circRNA that is 304nt in spliced sequence length, and its gene is located at chr20: 47570092–47580435. It derives from exons 6 and 7 of the ADP ribosylation factor guanine nucleotide exchange factor 2 (ARFGEF2) gene. Therefore, we determined the relationship between the clinicopathological features or prognosis of HCC.
Patients and methods
Patients and samples
Two hundred paired HCC and adjacent noncancerous tissues were collected from Tongren Hospital, Shanghai Jiao Tong University School of Medicine, People's Republic of China from August 2013 to September 2015. At the same time, we collected peripheral blood samples from 100 HCC patients, 50 hepatitis B patients and 50 healthy controls. The blood samples of HCC were collected before surgery and after surgery (more than 21 days after surgery), respectively. Five milliliter blood was obtained from each subject in K2-EDTA tubes and centrifuged at 1600g for 10 mins at 4°C, followed by 12,000g for 10 mins at 4°C. The supernatants (plasma) were collected and stored at −80°C until use. HCC was verified by pathologists for routine diagnosis. The clinical staging was determined by the Barcelona clinic liver cancer staging system. The patients underwent preoperative chemoradiotherapy were excluded. All specimens were stored at −80°C in RNA fixer reagent till use. The study was approved by the Human Ethics Committee of Tongren Hospital, Shanghai Jiao Tong University School of Medicine. The study complied with Declaration of Helsinki and all patients agreed to participate in the study and gave written informed consent.
Cell culture
The HCC cell lines HepG2, HuH7, MHCC97H, PLC/PRF/5 as well as the immortalized human hepatic cell line L02 were obtained from ATCC. Cells were cultured in DMEM (HyClone, USA) containing 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Total RNA extraction and reverse transcription
Total RNA from tissues was extracted by using the Trizol reagent (Invitrogen, USA) according to the instruction from the manufacturer. Total RNA from plasma specimens was extracted by using miRNeasy Mini Kit (Qiagen, German). RNA concentration was measured with a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using PrimeScriptTM RT Master Mix (TaKaRa, Dalian, People’s Republic of China) according to the manufacturer’s protocol (with random primers).
qRT-PCR
qRT-PCR was performed with SYBR Premix Ex TaqTM II (TaKaRa, Dalian, People’s Republic of China) on LightCycler 480II real-time PCR system (Roche) following manufacturer’s instructions. The primers used for the PCR reactions were hsa_circ_0003998 (forward: 5ʹ-CAGGAGGTGGTGAAGGACAT-3ʹ and reverse: 5ʹ-CCTGACTGTGCTTCAAACGA-3ʹ) and GAPDH (forward: 5ʹ- GGGAGCCAAAAGGGTCAT-3ʹ and reverse: 5ʹ-GAGTCCTTCCACGATACCAA-3ʹ). These primers were synthesized by Sangon Biotech (Shanghai, People’s Republic of China). The data were analyzed using the ΔCt method. All results were expressed as the mean±SD from three repeated experiments.
RNA-seq
RNA-seq libraries were constructed based on the Illumina standard protocols. Total RNA was treated with RiboMinus Eukaryote Kit (Qiagen, Valencia, CA, USA) to remove ribosomal RNAs, and digested with RNAse R to remove linear RNAs. The enriched circRNAs were prepared by the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Beverly, MA, USA) following the Illumina protocol. The main steps involved: the fragmentation of circRNA, second-strand cDNA synthesis, end repair, add an A at the 3ʹ end and ligated to adaptors. The ligated cDNA products were purified and subjected to PCR amplification. The libraries were sequenced on the Illumina HiSeq2500 platform (Illumina,USA) on a 100 bp paired-end run.
Sanger sequencing
qRT-PCR products were inserted into a T vector for Sanger sequencing by determining its full-length by Sangon Biotech (Shanghai, People’s Republic of China).
Statistical analyses
All statistical data were analyzed using SPSS 13.0 (SPSS Inc., USA) and GraphPad Prism 5.0 (GraphPad software, USA). The differences in the levels of hsa_circ_0003998 were analyzed by the Student’s t-test for paired data. Independent sample t-test and one-way (ANOVA were determined correlations between expression levels of hsa_circ_0003998 and clinicopathological factors. The receiver operating characteristic (ROC) curve was generated to evaluate its diagnostic value. The cutoff value was calculated according to Youden’s index. Kaplan–Meier analysis with log-rank statistic was used to analyze the overall survival (OS). Independent prognostic factors were performed using the Cox regression model. Two-sided P<0.05 was considered statistically significant.
Results
Overview of circRNA profiles in HCC tissues
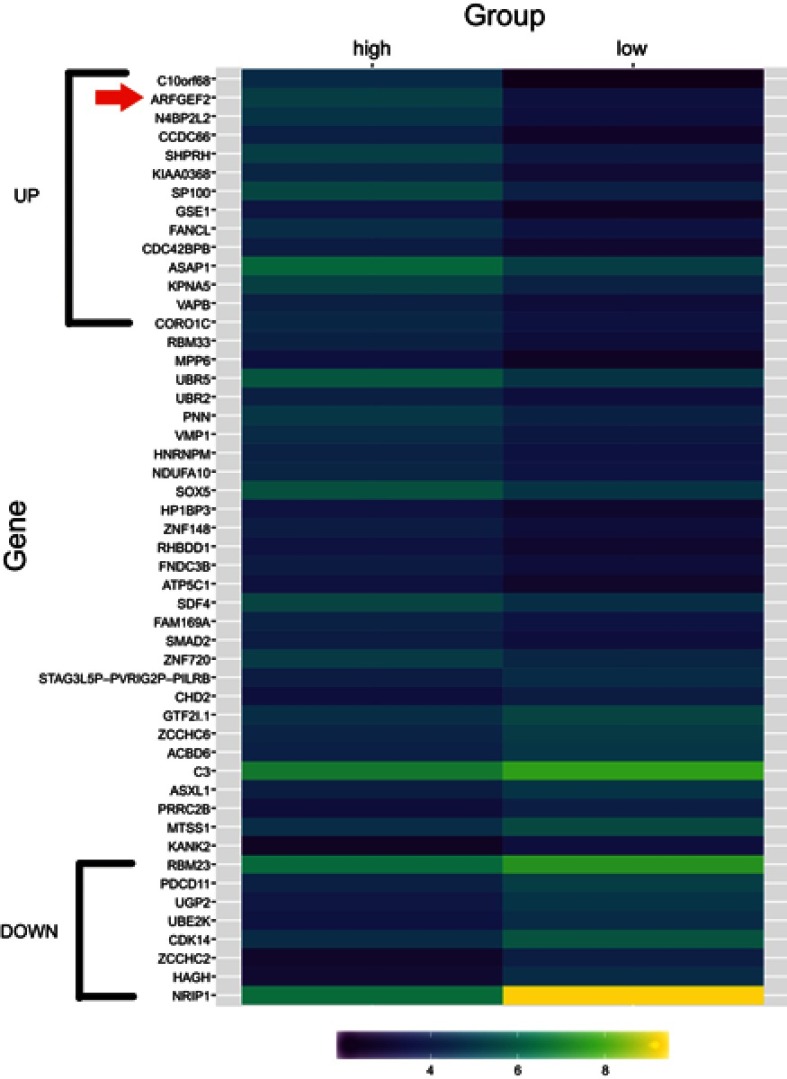
It was obtained that circRNA profiles from six cases of HCC tissues (three cases with or without portal vein invasion) (Table S1). Hierarchical clustering demonstrated the distributions of circRNA profiles (Figure 1). Overall, 121 circRNAs were co-expressed, including 14 upregulated and 8 downregulated circRNAs in HCC tissues with portal vein invasion compared to those without portal vein invasion. Among them, the most prominent circRNA was hsa_circ_0003998.
Table S1.
The patient profiles of HCC samples for RNA-sequencing
| Characteristics | Portal vein invasion (n=3) | Without portal vein invasion (n=3) |
|---|---|---|
| Gender | ||
| Male | 2 | 2 |
| Female | 1 | 1 |
| Age (years) | ||
| ≤60 | 3 | 3 |
| >60 | 0 | 0 |
| HCC history | ||
| Negative | 3 | 3 |
| Positive | 0 | 0 |
| HBsAg | ||
| Negative | 0 | 1 |
| Positive | 3 | 2 |
| Anti-HCV | ||
| Negative | 3 | 3 |
| Positive | 0 | 0 |
| AFP | ||
| ≥400 ng/mL | 3 | 3 |
| <400 ng/mL | 0 | 0 |
| Child-Pugh classification | ||
| Child-Pugh A | 1 | 1 |
| Child-Pugh B | 2 | 2 |
| Tumor diameter | ||
| ≥5 cm | 2 | 2 |
| <5 cm | 1 | 1 |
| TNM | ||
| I/II | 1 | 1 |
| III/IV | 2 | 2 |
| Differentiation | ||
| High/moderate | 1 | 2 |
| Low | 2 | 1 |
| BCLC stage | ||
| Stage A | 0 | 0 |
| Stage B | 0 | 2 |
| Stage C | 3 | 1 |
Abbreviations: HCC, hepatocellular carcinoma; HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; BCLC, Barcelona clinic liver cancer; AFP, alpha-fetoprotien.
Figure 1.
Overview of the RNA-seq signatures.
Notes: Hierarchical clustering analysis showed distinguishable circRNA profiles among HCC tissues with or without portal vein invasion (high: with portal vein invasion, low: without portal vein invasion). Yellow indicated higher expression level and purple indicated lower expression level. The red arrow stands for hsa_circ_0003998.
Abbreviation: HCC, hepatocellular carcinoma.
Hsa_circ_0003998 was up-regulated in HCC tissues and cells
It was measured the expression of hsa_circ_0003998 in 200 paired human HCC and adjacent noncancerous tissues and HCC cell lines using qRT-PCR. The specificity of products was confirmed using electrophoresis (Figure 2A). The accuracy was confirmed by Sanger sequencing (Figure 2B). Hsa_circ_0003998 was up-regulated in HCC tissues (P<0.001) (Figure 2C) and HCC cell lines (HepG2, HuH7, MHCC97H) (P<0.001) (Figure 2D) .
Figure 2.
hsa_circ_0003998 expression features. (A) Electrophoresis of qRT-PCR products. Lane M is DL2000 Marker, lanes 1–5 are products. (B) Validation of splicing junction site through sequencing of qRT-PCR products. (C) High levels of hsa_circ_0003998 in HCC tissues (n=200, P<0.001). (D) High levels of hsa_circ_0003998 in HCC cell lines. The △Ct value was determined by subtracting the Ct value of GAPDH from the Ct value of hsa_circ_0003998. Larger ΔCt value indicated lower level. **P<0.001 represented the statistical differences.
Correlations between hsa_circ_0003998 levels and clinicopathological factors in HCC patients
It was analyzed that the association between hsa_circ_0003998 levels and clinicopathological factors of HCC patients. As shown in Table 1, upregulation of hsa_circ_0003998 level was significantly correlated with higher serum AFP level (P=0.003), larger tumor diameter (P=0.009), microvascular invasion (MVI) (P=0.028) and lower differentiation level (P=0.023).
Table 1.
The relationship between hsa_circ_0003998 expression levels (△Ct) and clinical characteristics of HCC patients
| Characteristics | Patient number (total=200) | Mean±SD | P-value |
|---|---|---|---|
| Gender | |||
| Male | 108 | 7.6±1.59 | 0.077 |
| Female | 92 | 8.01±1.68 | |
| Age (years) | |||
| ≤60 | 91 | 7.73±1.63 | 0.678 |
| >60 | 109 | 7.83±1.66 | |
| HCC history | |||
| Negative | 176 | 7.82±1.84 | 0.474 |
| Positive | 24 | 7.56±1.84 | |
| Smoking | |||
| Negative | 140 | 7.85±1.57 | 0.384 |
| Positive | 60 | 7.63±1.8 | |
| Alcoholism | |||
| Negative | 134 | 7.89±1.52 | 0.213 |
| Positive | 66 | 7.56±1.87 | |
| HBsAg | |||
| Negative | 52 | 8.08±1.63 | 0.135 |
| Positive | 148 | 7.68±1.64 | |
| Anti-HCV | |||
| Negative | 194 | 7.8±1.66 | 0.368 |
| Positive | 6 | 7.19±1.13 | |
| AFP | |||
| ≥400 ng/mL | 132 | 7.55±1.7 | 0.003 |
| <400 ng/mL | 68 | 8.24±1.44 | |
| Child-Pugh classification | |||
| Child-Pugh A | 168 | 7.82±1.65 | 0.528 |
| Child-Pugh B | 32 | 7.62±1.6 | |
| Tumor diameter | |||
| ≥5 cm | 142 | 7.61±1.72 | 0.009 |
| <5 cm | 58 | 8.22±1.36 | |
| TNM | |||
| I/II | 131 | 7.92±1.56 | 0.14 |
| III/IV | 69 | 7.54±1.78 | |
| MVI | |||
| Positive | 48 | 7.33±1.8 | 0.028 |
| Negative | 152 | 7.93±1.57 | |
| Differentiation | |||
| High/moderate | 135 | 7.98±1.48 | 0.023 |
| Low | 65 | 7.37±1.89 | |
| BCLC stage | |||
| Stage A | 102 | 7.76±1.75 | 0.95 |
| Stage B | 62 | 7.84±1.42 | |
| Stage C | 36 | 7.76±1.73 | |
| ALT | |||
| ≥50 U/L | 75 | 7.74±1.66 | 0.766 |
| <50 U/L | 125 | 7.81±1.64 | |
| AST | |||
| ≥40 U/L | 60 | 7.73±1.79 | 0.748 |
| <40 U/L | 140 | 7.81±1.58 | |
| Liver cirrhosis | |||
| Yes | 122 | 7.66±1.71 | 0.191 |
| No | 78 | 7.98±1.52 |
Note: Bold values indicate P<0.05.
Abbreviations: HCC, hepatocellular carcinoma; HBsAg, hepatitis B surface antigen; HCV: hepatitis C virus; AFP, alpha-fetoprotien; MVI, microvascular invasion; BCLC, Barcelona clinic liver cancer; ALT, alanine aminotransferase; AST, aspartate transaminase.
Potential diagnostic values of hsa_circ_0003998
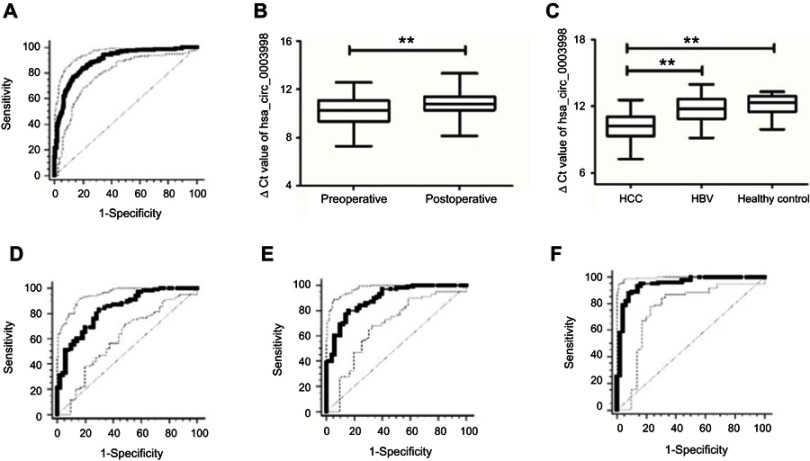
ROC curves were used to determine the diagnostic value of hsa_circ_0003998 in distinguishing HCC from adjacent noncancerous tissues. It was observed that the area under the ROC curve (AUC) was 0.894 (95% CI=0.86–0.922, P<0.001, Figure 3A). The sensitivity and specificity were 0.84 and 0.8, respectively. The value of the cutoff was 9.205.
Figure 3.
hsa_circ_0003998 as a biomarker. (A) ROC curves of hsa_circ_0003998 for discriminating between HCC and noncancerous tissues (P<0.001). (B) The plasma levels of hsa_circ_0003998 in preoperative and postoperative patients (P<0.001). (C) The plasma levels of hsa_circ_0003998 in HCC, hepatitis B patients and healthy controls (P<0.001). (D) ROC curves of hsa_circ_0003998 for discriminating between HCC and hepatitis B patients (P<0.001). (E) ROC curves of hsa_circ_0003998 for discriminating between HCC and healthy controls (P<0.001). (F) ROC curves of combination of hsa_circ_0003998 and AFP for discriminating between HCC and healthy controls (P<0.001). **P<0.001 represented the statistical differences.
Abbreviations: ROC, receiver operating characteristic; HCC, hepatocellular carcinoma; AFP, alpha-fetoprotien.
The plasma level of hsa_circ_0003998 in postoperative patients was significantly decreased than those in preoperative patients (post vs pre: 10.79±0.97 vs 10.21±1.2, P<0.001) (Figure 3B). The plasma level of hsa_circ_0003998 in HCC patients was significantly higher than those in hepatitis B patients (P<0.001) and healthy controls (P<0.001) (Figure 3C). ROC curve showed that, comparing with hepatitis B patients and healthy controls, hsa_circ_0003998, respectively, had an AUC value of 0.833 (95% CI=0.763–0.889, P<0.001) (Figure 3D) and 0.892 (95% CI=0.831–0.937, P<0.001) (Figure 3E). Besides, their sensitivity and specificity were 0.83, 0.7 and 0.8, 0.84, respectively. The values of cutoff were 11.345 and 11.255, respectively. Moreover, the combination of hsa_circ_0003998 and AFP for discriminating between HCC and healthy controls showed the highest AUC value of 0.947 (95% CI=0.898–0.977, P<0.001)(Figure 3F). The sensitivity and specificity were 0.88 and 0.92, respectively.
Potential prognostic values of hsa_circ_0003998
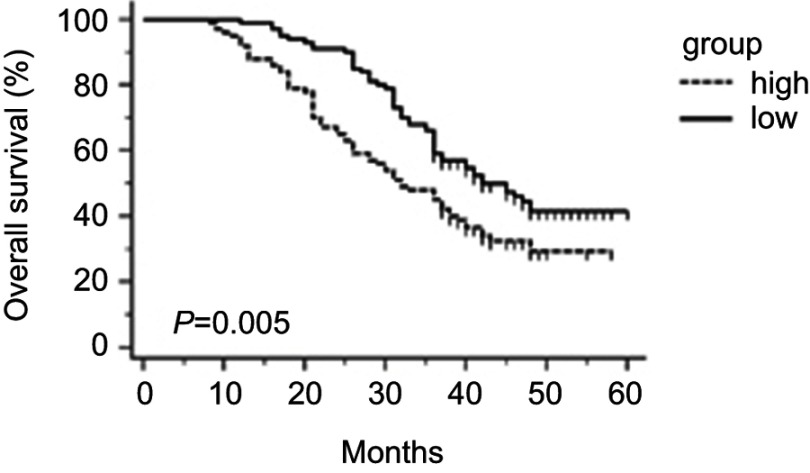
The HCC patients were divided into high (n=100) and low (n=100) expression groups according to the median level of hsa_circ_0003998 (ΔCt=8.235) in HCC tissues. Kaplan–Meier survival curves revealed that high hsa_circ_0003998 expression was significantly correlated with poor OS(P=0.005, Figure 4).
Figure 4.
The overall survival analysis in HCC patients by Kaplan–Meier method (P=0.005).
Abbreviation: HCC, hepatocellular carcinoma.
On univariate analysis, AFP (P=0.013), TNM stage (P=0.044) and hsa_circ_0003998 expression (P=0.006) were correlated with poor OS in HCC patients (Table 2). Furthermore, Cox multivariate analysis revealed that hsa_circ_0003998 (P=0.003) and AFP (P=0.008) were the independent prognostic factors for poor OS (Table 2).
Table 2.
Univariate and multivariate analyses of various prognostic parameters in HCC
| Parameters | Regression coefficient β | HR (95% CI) | P-value |
|---|---|---|---|
| Univariate analysis | |||
| Gender | −0.309 | 0.734 (0.513–1.05) | 0.091 |
| Age | −0.026 | 0.974 (0.682–1.391) | 0.885 |
| Smoking | 0.212 | 1.237 (0.845–1.81) | 0.274 |
| Alcoholism | 0.025 | 1.025 (0.702–1.496) | 0.898 |
| HBsAg | −0.003 | 0.997 (0.668–1.486) | 0.997 |
| AFP | 0.5 | 1.648 (1.112–2.444) | 0.013 |
| Tumor diameter | 0.286 | 1.331 (0.889–1.993) | 0.165 |
| TNM | 0.374 | 1.454 (1.01–2.093) | 0.044 |
| Differentiation | 0.306 | 1.359 (0.938–1.967) | 0.105 |
| BCLC stage | −0.039 | 0.962 (0.76–1.218) | 0.747 |
| ALT | 0.052 | 1.053 (0.732–1.514) | 0.781 |
| AST | 0.191 | 1.21 (0.827–1.771) | 0.326 |
| Liver cirrhosis | 0.232 | 1.261 (0.871–1.826) | 0.22 |
| Hsa_circ_0003998 expression | −0.505 | 0.604 (0.422–0.864) | 0.006 |
| Multivariate analysis | |||
| AFP | 0.538 | 1.713 (1.153–2.544) | 0.008 |
| Hsa_circ_0003998 expression | −0.539 | 0.583 (0.407–0.836) | 0.003 |
Note: Bold values indicate P<0.05.
Abbreviations: HCC, hepatocellular carcinoma; HBsAg, hepatitis B surface antigen; AFP, alpha-fetoprotien; BCLC, Barcelona clinic liver cancer; ALT, alanine aminotransferase; AST, aspartate transaminase; HR, hazard ratios; CI, confidence interval.
Discussion
HCC is the sixth high morbidity and the fourth leading cause of cancer mortality worldwide.2 HCC is accounted for 80–90% cases of liver cancer. It is rather distressed that the majority of HCC patients are diagnosed at advanced stages, missing the opportunity for curative treatment.18 Therefore, it is imperative to discover novel tumor markers for early diagnosis of HCC.
Recent reports revealed that the imbalanced expression of circRNAs is common in tumor and played crucial roles in biological processes.19 Moreover, it is suggested that circRNA might act as a potential diagnostic and prognostic biomarker in HCC.20,21 For example, it has been found that hsa_circ_0005075 has diagnostic value in HCC (AUC=0.94).22 Currently looking for a diagnostic target for HCC has become a hot spot for researchers Downregulation of hsa_circ_0001445 in HCC is correlated with tumor differentiation, tumor stage and MVI, and considered as a risk factor for OS and recurrence-free survival.23
In the current study, circRNA profiles screened by RNA-seq showed that hsa_circ_0003998 was significantly higher in HCC tissues with portal vein invasion than in HCC tissues without portal vein invasion. HCC has the characteristics of easy invasion and metastasis, which are the critical factors of high recurrence rate and poor prognosis.24 To explore the possible effect of hsa_circ_0003998 in HCC, we found that it was significantly higher in HCC tissues than in adjacent noncancerous tissues (P<0.001). Moreover, hsa_circ_0003998 level was significantly higher in HCC cell lines than in human hepatic cell line (P<0.001). Collectively, these findings demonstrated that hsa_circ_0003998 was significantly upregulated in HCC.
We also found that upregulation of hsa_circ_0003998 level was significantly associated with higher serum AFP level, larger tumor diameter, lower differentiation level and MVI. The plasma level of hsa_circ_0003998 in HCC patients was significantly higher than those in hepatitis B patients and healthy controls. Besides, the plasma level of hsa_circ_0003998 was significantly reduced after operation. The phenomenon might be due to the decreasing release of tumor-derived nucleic acid after tumor excision. The results suggest that hsa_circ_0003998 plays an important role during tumorigenesis of HCC.
The early diagnosis of HCC is of paramount importance for good prognosis. Currently, combination of radiology and serum AFP levels fail to diagnose HCC at the early stage.25 Therefore, it is urgent to recognize efficient molecular markers for HCC diagnosis. CircRNAs are endogenous, conserved, abundant and steady in cells, and are related to carcinomas.26 Moreover, in comparison to their cognate linear isoforms, the expression levels of circRNAs are above 10 fold.27 They were appealed as molecular biomarkers for cancer diagnosis and monitoring. The ROC analysis showed that AUC was 0.894, sensitivity and specificity were 0.84 and 0.8 for distinguishing HCC from adjacent noncancerous tissues. Comparing with healthy controls, hsa_circ_0003998 had an AUC value of 0.892, the sensitivity and specificity were 0.8 and 0.84, respectively. Moreover, the combination of hsa_circ_0003998 and AFP showed the highest AUC value of 0.947, the sensitivity and specificity were 0.88 and 0.92, respectively. Thus, hsa_circ_0003998 was recognized as a novel biomarker for distinguishing HCC from non-HCC with favorable sensitivity and specificity.
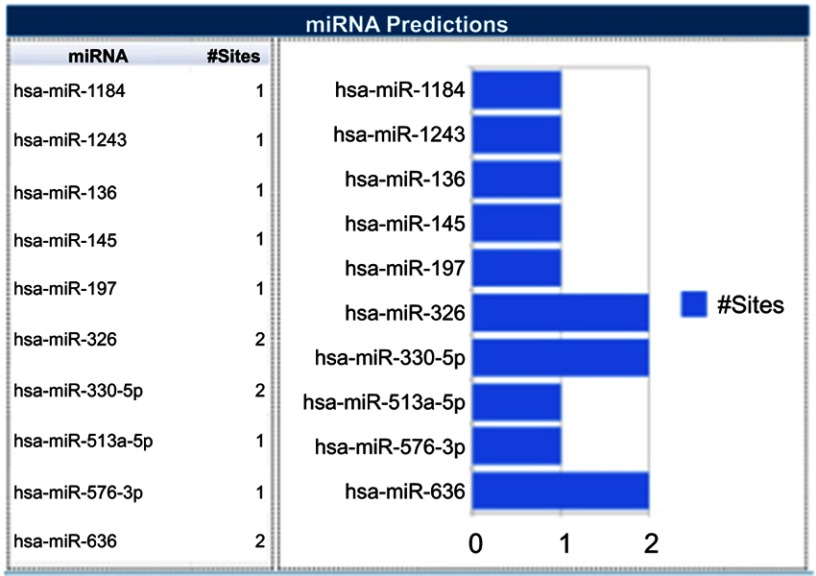
KaplanMeier analysis suggests that HCC patients with high level of hsa_circ_0003998 had significantly shorter OS than those with low level. Moreover, the Cox regression suggests that hsa_circ_0003998 was the independent favorable predictor of OS in HCC patients, but the functions of circRNAs in carcinogenesis and tumor progression remain to be elucidated clearly. Increasing evidence suggests that circRNA regulates tumor progression via acting as a miRNA sponge or regulating the expression of parental gene.9,28,29 This is supported by the finding that circ-ITCH could function as miR-17 and miR-224 sponges and regulate p21 and PTEN genes to suppress aggressive biological behaviors of bladder cancer.30 CircMTO1 may also sponge miR-9 and inhibit its oncogenic effect with circMTO1/miR-9/p21 axis in HCC.16 Using bioinformatics program (Circular RNA Interactome, https://circinteractome.nia.nih.gov/index.html), it is discovered that miR-1184, miR-1243, miR-136, miR-145, miR-197, miR-326, miR-330-5p, miR-513a-5p, miR-576-3p and miR-636 are the potential binding targets of hsa_circ_0003998 (Figure 5). This is in line with the findings form Yu et al31 that hsa_circ_0003998 acts as an oncogene in non-small cell lung cancer by modulating miR-326/Notch1. Such data suggest that hsa_circ_0003998 plays an important role through sponging miRNA in HCC, which may provide a direction for future study to explore the precise mechanism of hsa_circ_0003998 in carcinogenesis of HCC both in vivo and in vitro.
Figure 5.
TargetScan miRNA predictions.
Conclusion
In summary, hsa_circ_0003998 was expressed higher in HCC compared to the non-HCC. Hsa_circ_0003998 correlated with AFP, tumor diameter, differentiation and MVI might be used as a novel potential biomarker for early diagnosis and prognosis of HCC patients.
Acknowledgments
This work was supported by grants from Shanghai Tongren Hospital (numbers TRYJ201514 and 2019shtrxx09), the Project of Shanghai Municipal Commission of Health and Family Planning (numbers 20174Y0231 and 201640275), the National Natural Science Foundation of China (number 81672335).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary material
References
- 1.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Guo X, Lv X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. 2017;8(27):44050–44058. doi: 10.18632/oncotarget.17211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauzay C, Petit A, Bourgeois AM, et al. Alpha-foetoprotein (AFP): a multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39–44. doi: 10.1016/j.cca.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 6.Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Xie Q, He D, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18(1):834. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8(7):1142–1156. [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 10.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859(10):1245–1251. doi: 10.1016/j.bbagrm.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Zhang C, Lin J, Song X, Wang H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1275–1283. doi: 10.2147/CMAR.S166740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. 2017;16(10):3891–3902. doi: 10.1021/acs.jproteome.7b00519 [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036 [DOI] [PubMed] [Google Scholar]
- 16.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 17.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 18.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8(35):58405–58416. doi: 10.18632/oncotarget.16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332. doi: 10.3389/fonc.2018.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Zhou H, Jing W, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8). doi: 10.3390/cancers10110400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95(22):e3811. doi: 10.1097/MD.0000000000004864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 24.Yao T, Chen Q, Shao Z, Song Z, Fu L, Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;e22572. doi: 10.1002/jcla.22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta M, Gabriel H, Miller FH. Role of imaging in surveillance and diagnosis of hepatocellular carcinoma. Gastroenterol Clin North Am. 2018;47(3):585–602. doi: 10.1016/j.gtc.2018.04.013 [DOI] [PubMed] [Google Scholar]
- 26.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 29.Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5(3):317–333. doi: 10.1002/wrna.1213 [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. doi: 10.1186/s12943-018-0771-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, Jiang H, Zhang H, Li J. Hsa_circ_0003998 promotes cell proliferation and invasion by targeting miR-326 in non-small cell lung cancer. Onco Targets Ther. 2018;11:5569–5577. doi: 10.2147/OTT.S174750 [DOI] [PMC free article] [PubMed] [Google Scholar]