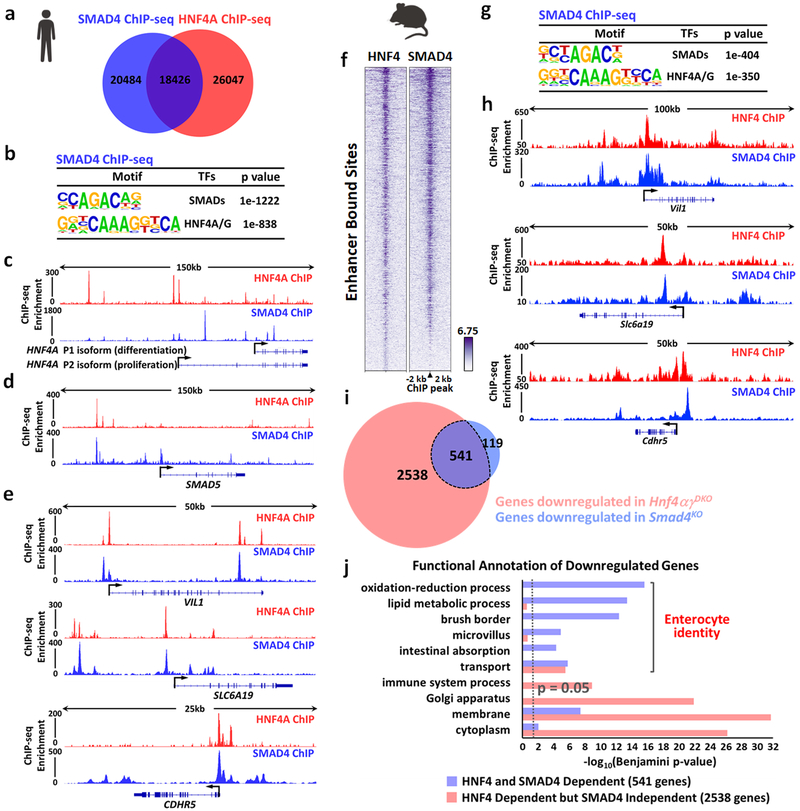
Figure 4. HNF4 and SMAD4 co-bind genomic enhancers, activating enterocyte-identity genes.
One replicate of human Caco-2 cell line and two replicates of biologically independent mouse tissues were used in ChIP-seq analysis. Statistical tests were embedded in MACS and HOMER. ChIP-seq in Caco-2 cells reveals that SMAD4 and HNF4A co-bind genomic regions, as evidenced by (a) Venn diagram (MACS P ≤ 10−5) and (b) HOMER motif analysis (MACS P ≤ 10−10). HNF4A and SMAD4 bind to gene loci of (c) HNF4A, (d) SMAD5 and (e) enterocyte differentiation genes (VIL1, SLC6A19 and CDHR5) in Caco-2 cells. ChIP-seq in mouse intestinal epithelium also reveals that SMAD4 and HNF4 co-bind genomic regions (n = 2 biologically independent mice each). f, Heatmap shows a similar binding pattern of SMAD4 (MACS P ≤ 10−5) and HNF4 (MACS P ≤ 10−3) at their enhancer bound sites. g, HOMER motif analysis of mouse SMAD4 ChIP-seq (MACS P ≤ 10−5, enhancer sites). h, HNF4 and SMAD4 co-bind to enterocyte differentiation genes (Vil1, Slc6a19, and Cdhr5) in mouse intestinal epithelium. i, Venn diagram shows highly overlapping target genes with significant downregulation (FDR < 0.05) in the intestine of Hnf4αγDKO (red circle) and genes with significant downregulation (FDR < 0.05) in Smad4KO (blue circle). Significant genes were called via Cuffdiff. (n = 3 biologically independent mice). j, Enterocyte-related functions are enriched among HNF4-SMAD4 co-dependent genes (common downregulated genes, 541 genes, purple bar) compared to genes only downregulated in Hnf4αγDKO but not in Smad4KO (2,538 genes, red bar). This suggests the combined functions of HNF4 and SMAD4 are specifically driving the transcriptome towards enterocyte identity. Benjamini P values were calculated using DAVID.