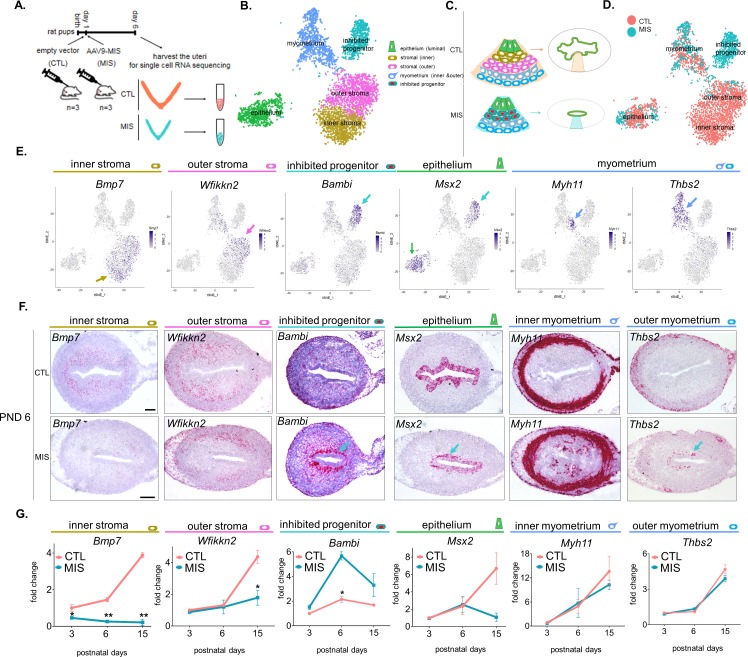
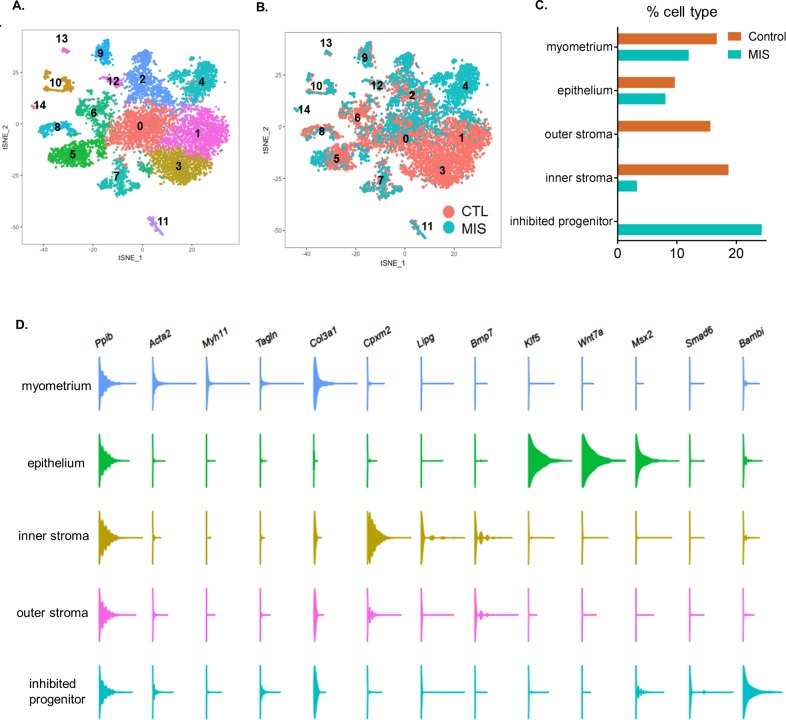
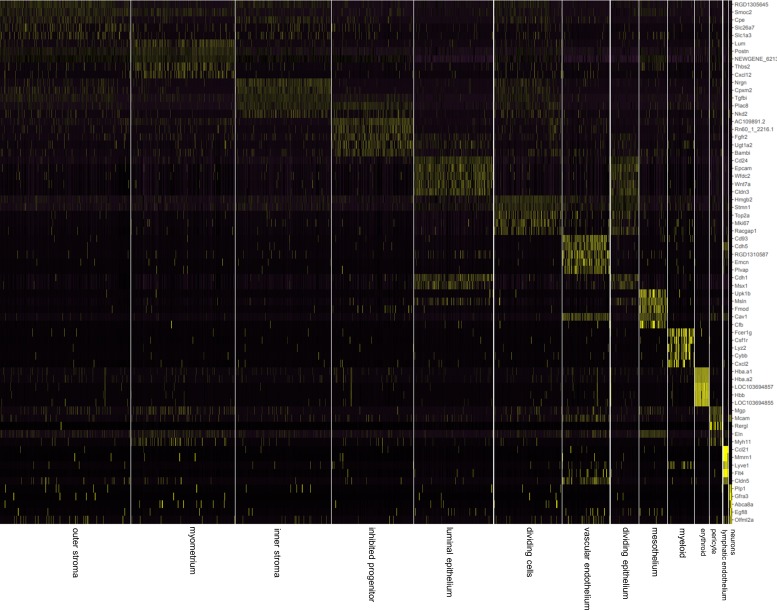
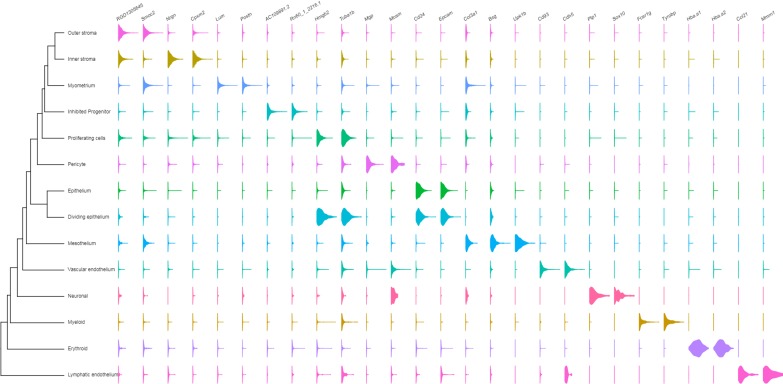
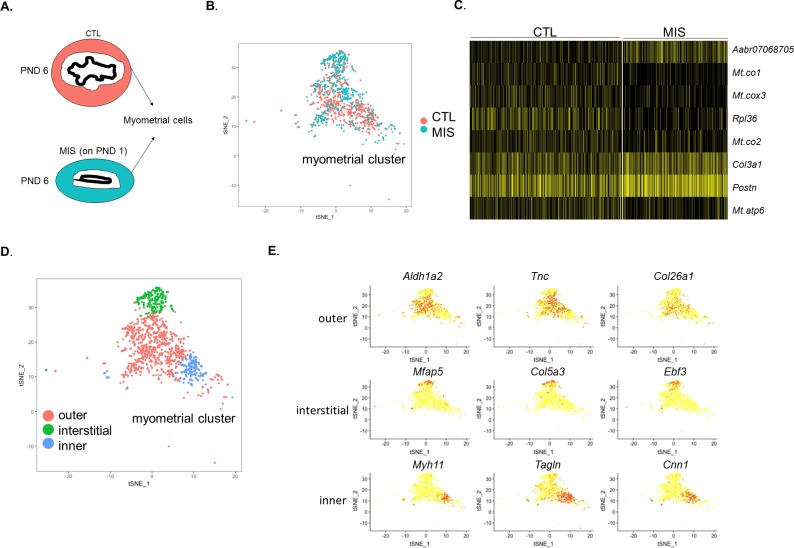
Figure 3. Single cell RNA sequencing of PND 6 uteri reveals distinct cell types and unique gene signatures in response to MIS treatment.
(A) Rat pups were treated with empty vector control (CTL) or AAV9-MIS (MIS) on PND 1, and euthanized on PND 6 (n = 3 for both). Following whole-tissue dissociation, RNA isolated from single uterine cells, were barcoded and sequenced using inDROP. (B) t-SNE plot of unbiased clustering of uterine cells, where each color-coded cluster represents one cell type/state (only the main uterine parenchymal clusters are represented) (Figure 3—source data 1). (C) Schematic representation of the differential cellular composition of control and MIS-treated uteri. (D) t-SNE plot of unbiased clustering of uterine cells (dots) color-coded by treatment with CTL (orange) and MIS (blue) (Figure 3—source data 1). (E) Gene expression levels of representative cell-type-specific markers for each cluster overlaid on t-SNE plots (featureplot, color-coded arrow refers to cell type). (F) RNAish stains of representative cell-type markers in transverse uterine sections of empty vector control (CTL) and AAV9-MIS (MIS) treated mice at PND 6. Scale bars = 100 µm, same for all sections. Mouse tissues were used for validation purposes as the RNA in situ probes were readily available for mice and the effect of MIS was conserved among mice and rats. (G) Rat pups treated with AAV9-MIS (MIS) or empty vector (CTL) on PND 1 were euthanized at different developmental time points (PND3, 6, and 15), and their uteri were harvested for QPCR analysis of one representative marker for each cluster (n > 2, unpaired Student’s t test, mean ± SEM, *(p<0.05), **(p<0.01)) (Figure 3—source data 3).