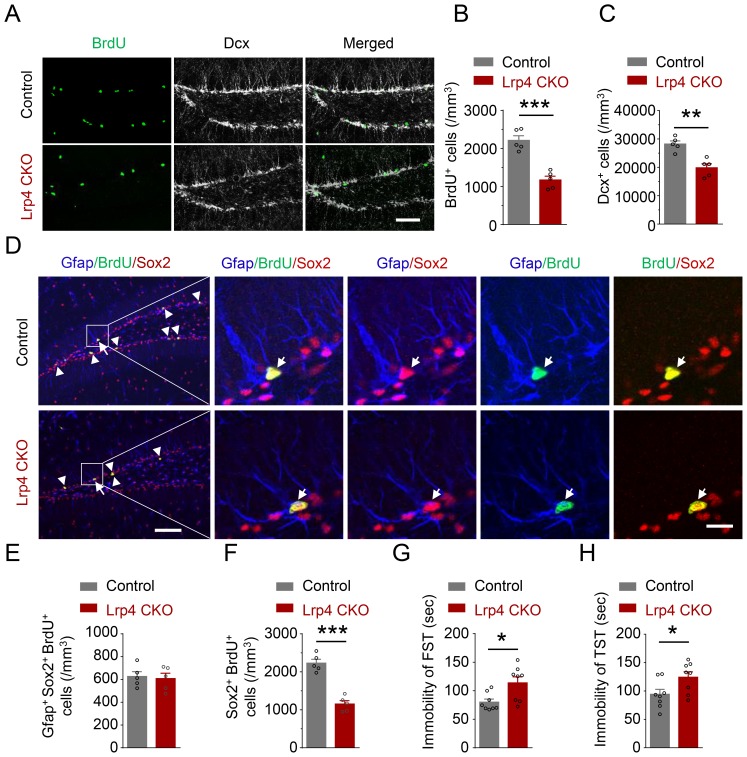
Figure 3. Reduced NSPCs proliferation and increased immobility of Lrp4 CKO mice.
(A–C) Reduced numbers of BrdU- and Dcx-labeled cells in Lrp4 CKO SGZ. (A) Representative images. Scale bar, 100 μm. (B–C) Stereological quantification of SGZ BrdU+ (B) and Dcx+ (C) cells. n = 5 for each group. Student’s t-test: t (8)=6.602, p=0.0002 for BrdU; t (8)=4.701, p=0.0015 for Dcx. (D–F) Reduced NPCs proliferation in Lrp4 CKO. (D) Representative images. The arrow indicated Gfap+/BrdU+/Sox2+, while the arrow head indicated BrdU+/Sox2+ cells. Scale bar, left 100 µm, right 20 µm. (E) Similar numbers of SGZ Gfap+Sox2+ BrdU+ NSCs between the two genotypes. n = 5 for each group. Student’s t-test: t (8)=0.2947, p=0.7757. (F) Decreased the density of Sox2+BrdU+ NPCs in Lrp4 CKO mice, compared with control. n = 5 for each group. Student’s t-test: t (8)=7.943, p<0.0001. (G–H) Increased duration of immobility in FST (G) and TST (H) of Lrp4 CKO mice, compared with control. n = 8 for each group. Student’s t-test: t (14)=2.826, p=0.0135 for FST; t (14)=2.332, p=0.0352 for TST. Data are mean ± s.e.m; *, p<0.05; **, p<0.01; ***, p<0.001.