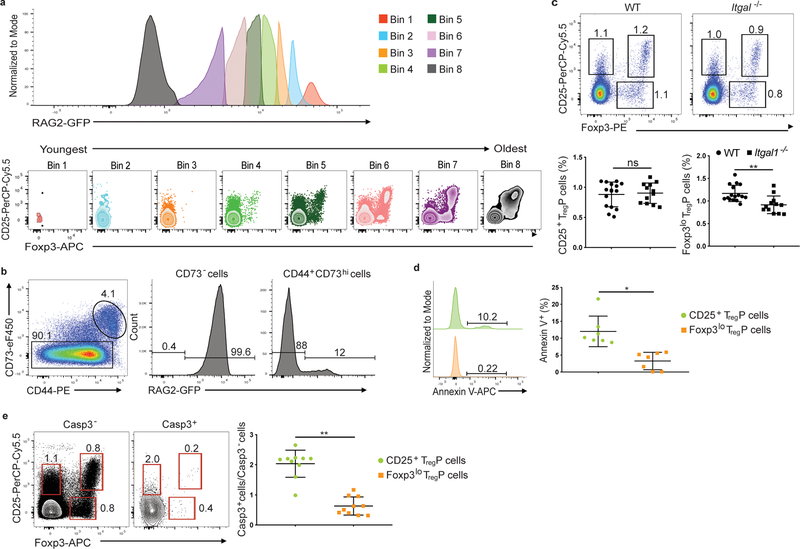
Figure 3.
CD25+ and Foxp3lo TregP cells are in discrete selection stages. a) Representative histograms of RAG2-GFP expression in CD4SP thymocytes obtained from thymi of RAG2-GFP mice. Bins are displayed from high (Bin 1) to low (Bin 8) RAG-GFP expression and cells in each bin are plotted for CD25 vs Foxp3 expression. Data shown are concatenated results from 3 mice and are representative of 7 independent experiments, n=9 mice. b) Representative flow cytometry plots of CD73 staining in CD4SP thymocytes and RAG2-GFP expression in CD73− and CD73+ compartments. Data is representative of 5 experiments, n=5 mice. c) Representative flow cytometry plots (top) and quantification of the percent of CD4+CD73− thymocytes differentiating into each TregP cell population in WT versus Itgal−/− thymi. Data represents 3 independent experiments, n= 16 WT and Itgal+/− mice, n=12 Itgal−/− mice. Data was analyzed by two-sided Mann-Whitney test. d) Left panels, Representative example of Annexin V expression on CD4+CD73−CD25+Foxp3− TregP (top green histogram) and CD4+CD73−CD25−Foxp3lo TregP (bottom orange histogram) thymocytes from Foxp3-GFP mice. Right panel, quantification of Annexin V staining on CD25+ TregP and Foxp3lo TregP. Data represents 3 independent experiments, n=9 mice. Data were analyzed by two-sided Wilcoxon matched-pairs signed rank test. e) Left and middle panels, represenative examples of staining for CD25 and Foxp3 on CD4+CD73− gated thymocytes from wild-type mice either negative (left panel) or positive (middle panel) for cleaved caspase-3. Right panel, quantification of the ratio of cleaved caspase-3-positive to cleaved caspase-3 negative CD25+ TregP (green circles) or Foxp3lo TregP; data represents 4 independent experiments, n=10 mice. Data were analyzed by two-sided Wilcoxon matched-pairs signed rank test. All data are displayed as mean ± SD. *P<0.05, **P<0.005, ns-not significant.