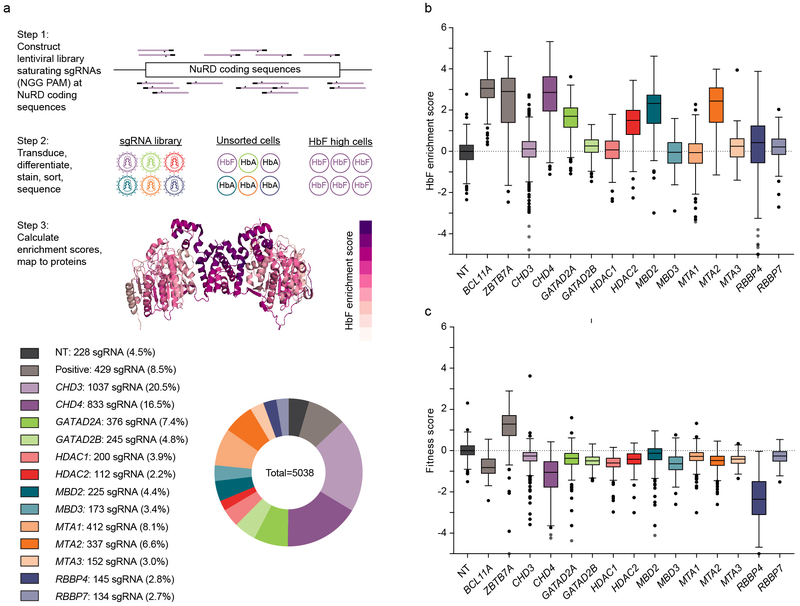
Fig. 1: Dense mutagenesis of NuRD genes by CRISPR-Cas9 pooled screening.
a, Upper panel shows overview of the CRISPR-Cas9 pooled lentiviral screen. All NGG PAM restricted sgRNAs targeting 13 NuRD genes plus BCL11A and ZBTB7A targeting and nontargeting sgRNAs synthesized by oligonucleotide array and cloned as pool to lentivirus. HUDEP-2 cells were transduced (n=3), selected, and subject to erythroid differentiation culture. Genomic DNA was isolated from unsorted and HbF+ cells. Integrated sgRNA libraries were amplified and deep sequenced to determine sgRNA representation. HbF enrichment scores calculated by comparing sgRNA abundance in HbF-high to unsorted cells, fitness score calculated by comparing sgRNA abundance in unsorted cells to library. Enrichment scores were mapped to linear and 3D protein maps. As an example, HbF enrichment scores from MTA2 and HDAC2 mapped to dimer of PDB ID 5ICN. Lower panel shows sgRNA library distribution. b, c, HbF enrichment (b) and fitness (c) scores for each sgRNA (sample size per lower panel a) targeting NuRD genes and controls. Boxplot shows median, 25th and 75th percentiles with whiskers and outliers per Tukey method.