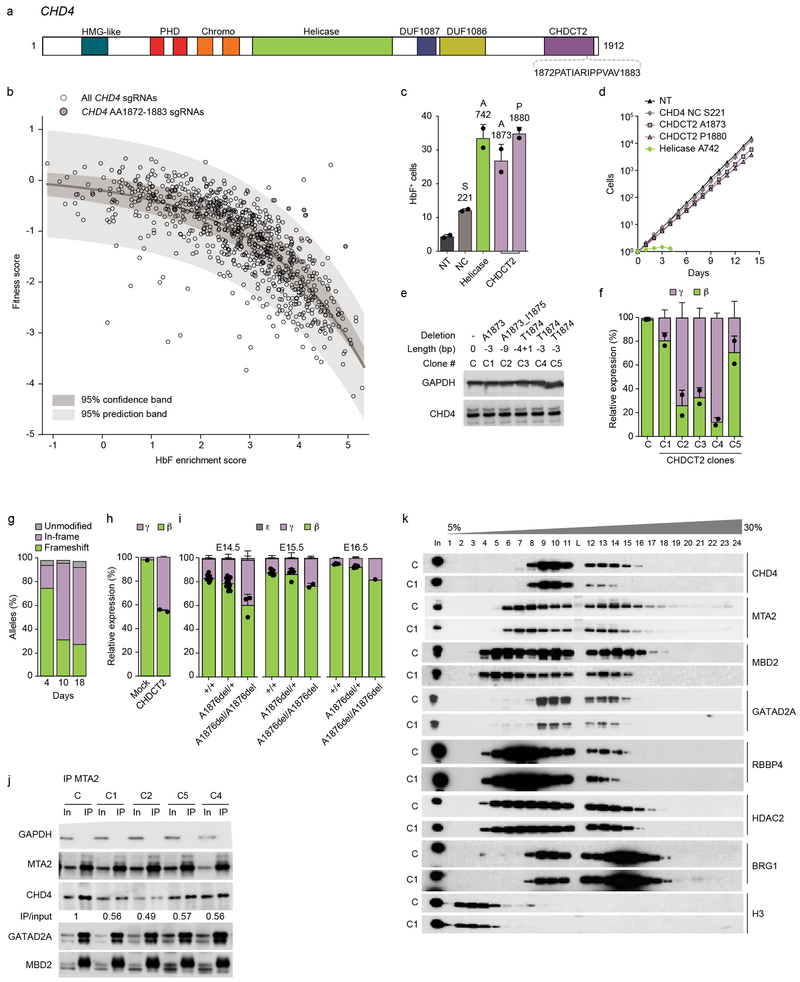
Fig. 5: Targeting CHD4 CHDCT2 uncouples HbF induction from cytotoxicity.
a, CHD4 domains. b, Dots indicate HbF enrichment and fitness scores of all 833 sgRNAs targeting CHD4. 95% confidence and prediction bands after non-linear least squares curve fitting. Purple dots show sgRNAs targetting AA1872-1883, the only nonoverlapping contiguous 12 AA segment of CHD4 significantly enriched in outliers from prediction band (p = 2.93 ×10−11, Fisher’s exact test, two-sided). sgRNA scores average of three experiments. c, Fraction HbF+ cells (%) following sgRNA targeting CHD4 NC, helicase, CHDCT2 domain or nontargeting (NT) (n = 2, error bars represent mean and SD). d, HUDEP-2 expansion following sgRNA targeting CHD4 S22, A742, A1873, P1880 and NT (average from three experiments). e, Immunoblot from CHD4 hemizygous in-frame deletion clones. Representative of three experiments. f, β- and γ-globin RT-qPCR in CHD4 hemizygous in-frame deletion clones (mean and SD from n = 2). g, CD34+ HSPCs electroporated with SpCas9:sgRNA targeting CHDCT2 A1873 with indels quantified by amplicon sequencing after 4, 10, or 18 day erythroid culture. h, β- and γ-globin expression in human erythroid precursors (mean and SD from n = 2). i, β-, ε- and γ-globin expression in fetal livers from E14.5, E15.5 and E16.5 Chd4A1876del/+;β-YAC+ × Chd4A1876del/+;β-YAC+ embryos (mean and SD, sample size per Supplementary Fig. 7d). j, IP MTA2 followed by NuRD subunit immunoblot. Quantification of CHD4 by IP/input ratio. k, Density sedimentation analyses of CHDCT2 domain hemizygous in-frame deletion HUDEP-2 clone. Each experiment (j-k) was repeated at least three times with similar results.