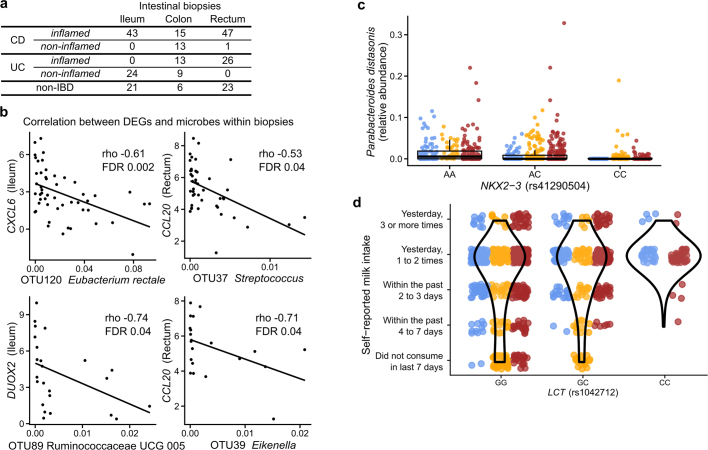
Extended Data Fig. 6. Mucosal communities and human genetics.
a, Number of biopsy samples available for each biopsy location and inflammation status. b, DEGs (Fig. 4a) with newly identified significant correlations with OTU abundances in biopsies (partial Spearman correlation conditioned on disease status, BMI, age at consent and sex; FDR P < 0.05; n = 54 in ileum and n = 52 independent 16S–RNA-seq pairs; full table in Supplementary Table 33). c, A limited subset of the microbiome trended with genetic variants in targeted testing, including the strongest trend shown here of Parabacteroides distasonis with genotypes of NKX2-3 (a known IBD-associated locus103; boxplots show median and lower/upper quartiles; whiskers show inner fences). This is the most significant association by P value among all tested associations between metagenomic taxa and five known IBD loci (nominal significance P = 0.006; no associations passed FDR P < 0.05, mixed effect model with age, sex, antibiotic and immunosuppressant use and first 20 genetic principal components as covariates while specifying subjects as random effects; Wald test; n = 84 subjects of European ancestry with exomes and 960 metagenomes; full results in Supplementary Table 34). d, Association between rs1042712 SNP in the LCT locus and self-reported milk intake from dietary recall. Self-reported short-term milk intake (from dietary recalls accompanying stool samples) was significantly associated with the count of C alleles (29.8% allele frequency) at rs1042712 in the LCT gene locus using a linear mixed effect model accounting for age, sex, first 20 genetic principal components and with subjects as random effects (P = 0.028, linear mixed effect regression with Wald test, see Methods). All available data are plotted for unique subjects of European ancestry with exome data (per-genotype subject count (GG/GC/CC):50/26/8). Differences between IBD and non-IBD groups are not statistically significant (odds ratio 0.27; 95% CI 0.05–1.33; P = 0.10; n = 84 subjects of European ancestry with exomes and 960 dietary surveys; model: IBD (yes|no) ~ intercept + SNP + sex + age + PC1–PC20).