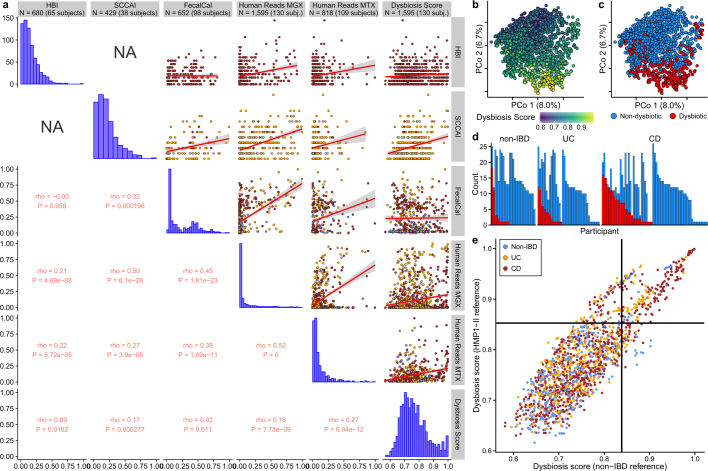
Extended Data Fig. 3. Patient-reported, molecular, and microbial disease activity measures.
a, Relationships between six measures of disease activity: the patient-reported HBI in CD, patient-reported SCCAI in UC, faecal calprotectin, the fractions of human reads from stool MGX and MTX, and a dysbiosis score defined here as departures from control population microbiome configurations (Fig. 2c). Rho values are Spearman correlations with ties broken randomly. Linear regression is shown (red line) with 95% confidence bound (shaded). Sample counts are presented in the title bar, though sample counts for a particular correlation may be less as samples must be paired. b, c, PCoA based on metagenomic species-level Bray–Curtis dissimilarities (n = 1,595 samples from 130 subjects), indicating dysbiosis score (b) and whether the sample was defined as dysbiotic (c). d, Number of dysbiotic samples per participant. Colour scheme as in c. e, Relationship between the dysbiosis score, when using the HMP1-II gut data set as reference (n = 553 from 249 subjects), compared to the non-IBD data set. The threshold for the dysbiosis classification is also shown (black lines). The two scores are highly correlated (Pearson ρ = 0.86; two-sided P < 2.2 × 10−16), as are the resulting dysbiosis classifications (odds ratio of 56; Fisher’s exact test P < 2.2 × 10−16). n = 1,595 samples from 130 subjects.