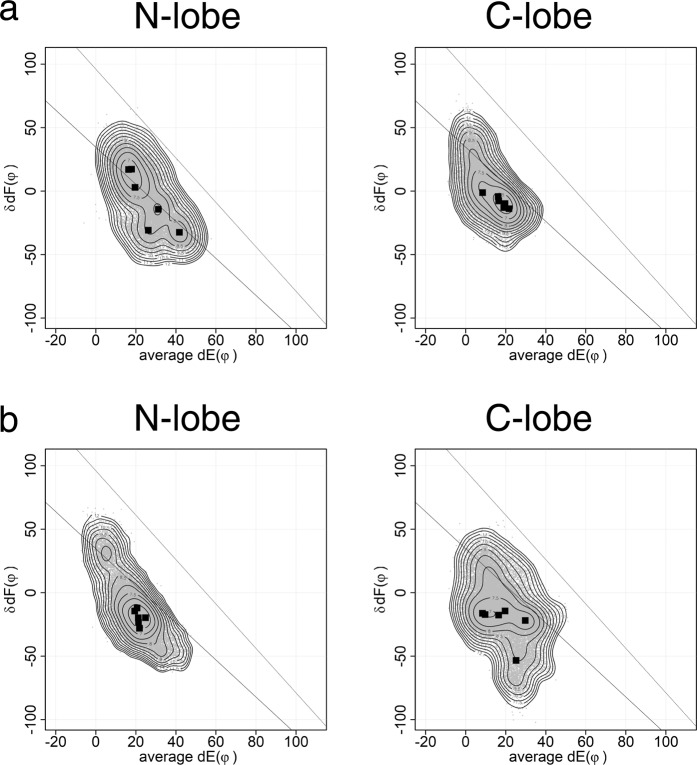
Figure 6.
The effects of amino acid sequence for conformational ensembles of each EF-lobe. (a) The amino acid sequences of N- and C-lobes in 1CLL were swapped. The model was built with SwissModel by homology modelling. (b) The amino acid sequence of the model used in (a) was reverted to the native sequence. The model was built with SWISS-MODEL by homology modelling using the structure used in (a) as a template. MD simulations were performed by using GROMACS 2016 on a Cray XC50. This figure summarizes six 100 ns MD results for each model. The dots are the conformations sampled at every 20 ps. Squares show the peak position for each MD run. The contour shows the free energy landscape of ensemble summed over six MD runs. The models generated by SWISS-MODEL are licensed under the CC BY-SA 4.0 Creative Commons Attribution-ShareAlike 4.0 International License.