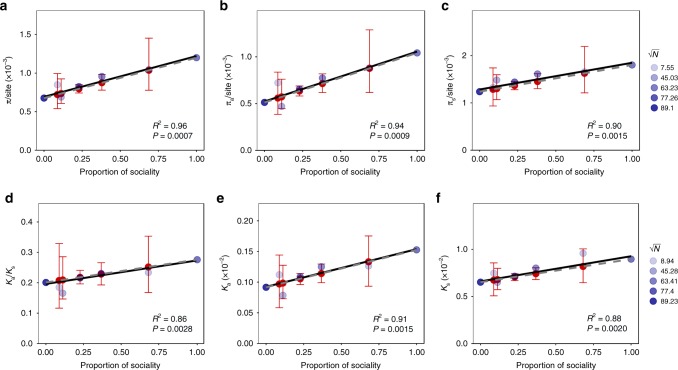
Fig. 4.
The impact of Red King processes on polymorphism and divergence. The top row of panels shows the relationship between nucleotide diversity in social genes at the full CDS a, nonsynonymous b, and synonymous sites c as a function of the proportion on conditionally expressed (sociality) genes within that group. Similarly, the bottom row of panels shows the relationship between the rates of nonsynonymous e, synonymous substitutions f, and the rate of protein evolution d in groups of genes as a function of the proportion of sociality genes in each group. In each panel the red points indicate the expected value for a group and the red error bars the 95% confidence interval (both calculated from a set of 10,000 permutations). The blue points indicate the observed values for each group of genes, with the darkness of the point being determined by the sample size (based on ). The dashed line indicates to the expected value of each parameter predicted from the sociality and non-sociality genes. The solid line corresponds to the best fit line from a weighted regression (where weightings are ) fitted to the observed values. Source data are provided as a Source Data file