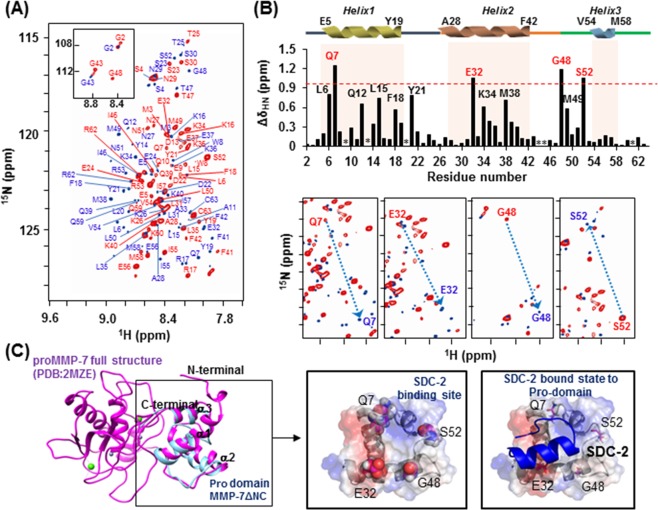
Figure 3.
Chemical shift perturbations of the 1HN and 15N resonances of proMMP-7 ΔNC upon binding the human syndecan-2 peptide ligand. (A) 1H-15N HSQC spectrum of proMMP-7 ΔNC in the absence and presence of S2-P. The spectra were collected at pH 6 using Bruker 850 and 900 MHz spectrometers, and NMR titrations were performed with 15N-labeled proMMP-7 ΔNC and S2-P at a molar ratio of 1:15. The backbone resonance assignments are shown in red (1:0) and blue (1:15). (B) The chemical shift perturbations of proMMP-7 ΔNC in the absence and presence of S2-P were assessed by overlapping the three titration points and performing interaction-site mapping of the proMMP-7/S2-P peptide complex using NMR titration (top). The average chemical-shift changes were calculated using the following formula: ΔδAV = [(Δδ1H)2 + (Δδ15N/5)2]1/2, where ΔδAV, Δδ1H, and Δδ15N represent the average chemical shift value, proton chemical shifts, and nitrogen chemical shift changes, respectively. The highly perturbed residues, Gln 7, Glu 32, Gly 48, and Ser 52, are marked in red. The chemical-shift movements of these residues were plotted in overlapped HSQC analyses (bottom). (C) The NMR structure of the pro-domain of human MMP-7 (PDB; 2MZE) (shown in magenta) and the modeled structure of proMMP-7 ΔNC (shown in cyan) were overlapped and visualized using a ribbon diagram (left). The side-chains of the four highly perturbed residues (Gln 7, Glu 32, Gly 48, and Ser 52) are shown as balls (center). HADDOCK model obtained for the proMMP-7/S2-P complex based on our NMR titration data, including the predicted binding site for S2-P on the pro-domain of proMMP-7 (right).