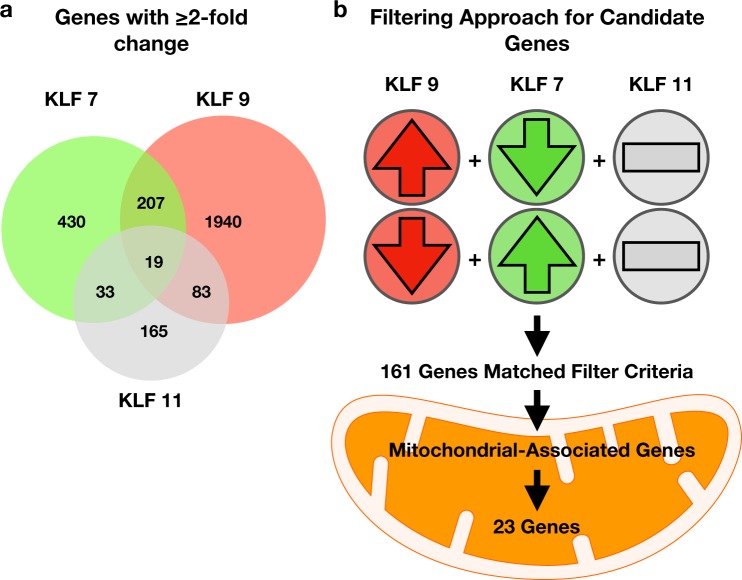
Figure 1.
Microarray data collected from KLF expression experiments in RGCs. (a) KLF regulated genes expressed greater than or equal to 2–fold in a positive or negative direction. Data collected from lentivirus mediated KLF7, -9 or -11 expression in cultured RGCs. (b) Filtering scheme to identify axon growth regulating genes from a list of 2 fold changing genes. Genes were grouped based on opposing outcomes of KLF expression on RGC axon growth enhancement/suppression. Arrows represent a directions fold change and bars represent no fold change. Up/down regulations were considered when changes were >2 fold with both KLFs, or slightly relaxed criteria of 2 fold change with one KLF and >1.5 with the other KLF. In all scenarios the KLF11 regulation of a particular gene had to be <1.5. A total of 163 genes met these criteria, of which 23 were considered mitochondrial associated based on gene ontology analysis. All fold changes were identified relative to lentivirus mCherry controls, and data were generated from N = 5 or more repeat RGC isolation, virus treatments, and microarray chips. Data analyzed with GenSpring 12.5–GX software, significantly expressed genes identified by a Oneway ANOVA and Welch test, p (Corr) cut-off of 0.05. See Supplementary Dataset 1, for raw, filtered, and candidate gene lists.