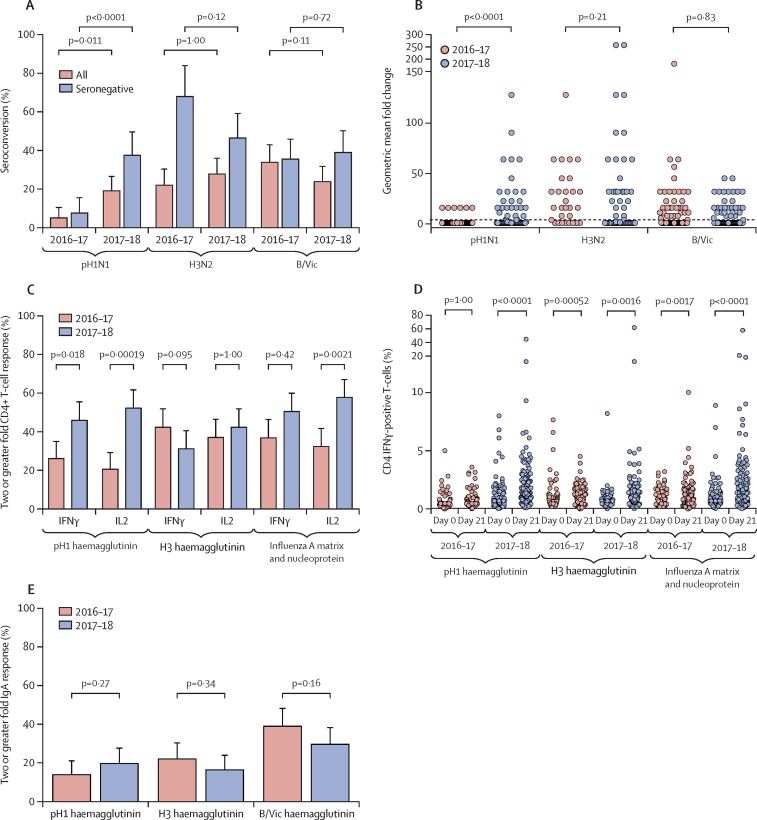
Figure 4.
Immunogenicity to pH1N1 with the 2016–17 and 2017–18 LAIV formulations
p values are Bonferroni-adjusted for multiplicity within each group of analyses. (A) Percentage of children seroconverting to each LAIV strain, comparing 2016–17 and 2017–18 formulations. Error bars represent the upper 95% CI. (B) Geometric mean fold change in serum haemagglutinin inhibition titre from baseline to day 21, comparing children seronegative at baseline given 2016–17 and 2017–18 LAIVs. Dotted line depicts a fold change of four. y axis is a logarithmic scale. (C) Influenza-specific CD4+ T-cell responses to vaccine strain-matched pH1 haemagglutinin (Cal09 in 2016–17 or NY15 in 2017–18), H3 haemagglutinin, influenza A matrix and nucleoprotein (both matched to LAIV backbone) peptide pools, comparing 2016–17 and 2017–18 LAIVs. Error bars represent the upper 95% CI. (D) Percentage of children with a twofold rise in influenza-specific CD4+ T-cell responses at day 21 after 2016–17 and 2017–18 LAIVs. y axis is a logarithmic scale. (E) Percentage of influenza-specific mucosal IgA responders given the 2016–17 and 2017–18 LAIVs. Error bars represent the upper 95% CI. pH1N1=pandemic H1N1. LAIV=live attenuated influenza vaccine. Cal09=A/17/California/2009/38. NY15=A/17/New York/15/5364. H3N2=A/17/Hong Kong/2014/8296. B/Vic=B/Texas/02/2013 (Victoria lineage). IFNγ=interferon γ.