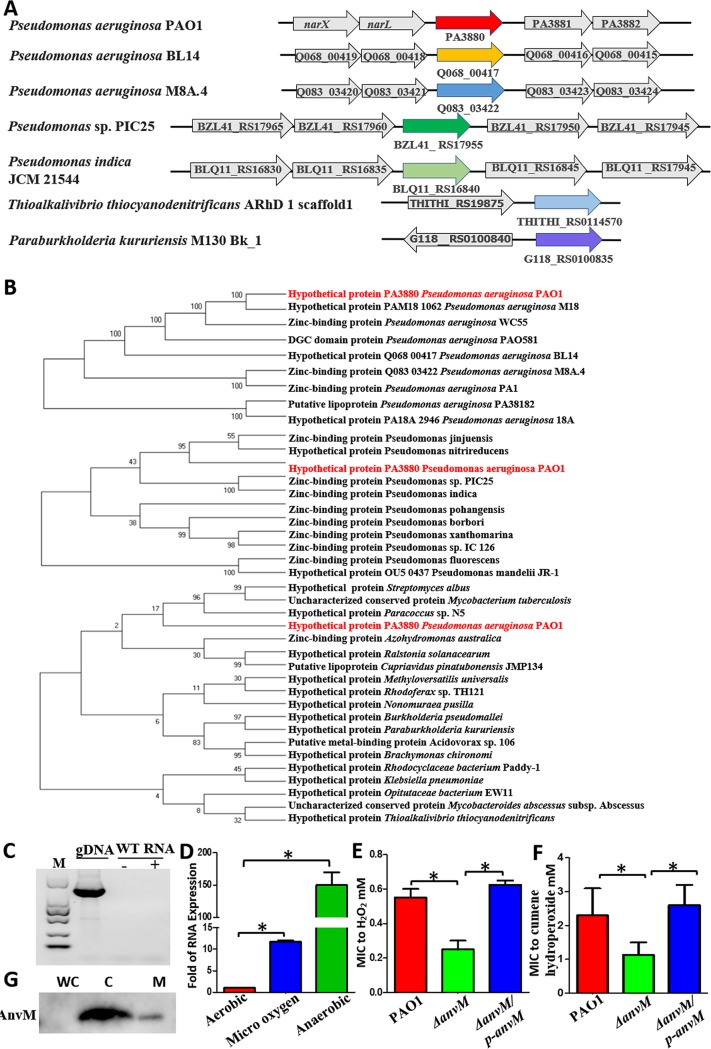
FIG 1.
Prevalence of AnvM-like protein. (A) The location of anvM in bacteria. (B) Phylogenetic tree of AnvM. Nine strains of P. aeruginosa, 11 strains of Pseudomonas spp., and 19 genera were included in the phylogenetic tree. The phylogenetic relations were inferred using the neighbor-joining method, and evolutionary distances were computed using the Poisson correction method. Analyses were performed with MEGA7 software. (C) RT-PCR showed that anvM was not in the same operon as narXL. gDNA, genomic DNA; M, molecular mass marker. (D) Real-time PCR showed that anvM is highly activated under microaerobic and anaerobic conditions. Anaerobic culturing was performed in 100-ml hermetically sealed bottles containing an AnaeroPack-Anaero (MGC), and micro-oxygen culturing was performed in 100-ml hermetically sealed bottles containing an AnaeroPack-MicroAero (MGC). (E and F) The anvM deletion strain was more sensitive to H2O2 (E) and cumene hydroperoxide (CHP) (F) than the WT strain. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way ANOVA with Tukey post hoc test). Serial 2-fold dilutions were prepared in LB that ranged from 10 mM to 0.005 mM H2O2 and 30 mM to 0.015 mM CHP. A 0.2-ml volume of each dilution was distributed over a 96-well polystyrene microwell plate. A 0.5 McFarland standard suspension (∼1.5 × 108 CFU/ml) was prepared from a fresh overnight culture of PAO1, strain ΔanvM, and strain ΔanvM/p-anvM, and 20 μl was added to each microwell in the 96-well plate in triplicate. Growth was monitored every 4 h for 24 h at 37°C using a Multiskan GO microplate spectrophotometer (Thermo Fisher Scientific, Vantaa, Finland). (G) Subcellular localization of AnvM. Western blot analysis showed that AnvM was located mostly on the cytoplasm (C) fraction and that a few AnvM proteins were located on the membrane (M) fraction of P. aeruginosa. The whole-cell lysates (WC) were used as controls.