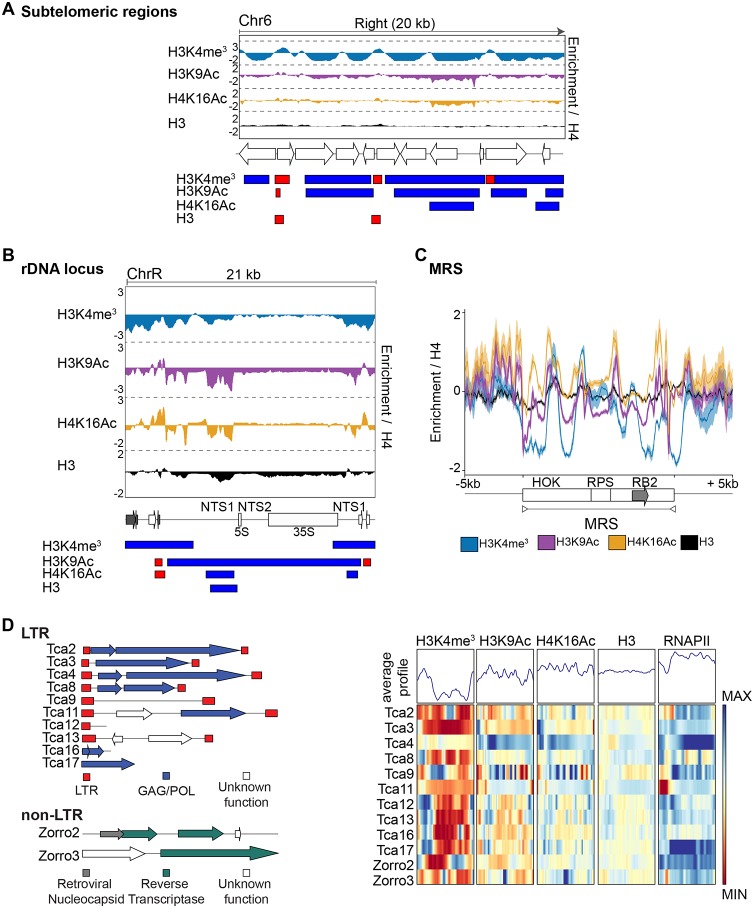
FIG 3.
Chromatin signature of C. albicans repetitive elements. (A) (Top) Fold enrichment (log2) of H3K4me3, H3K9Ac, H4K16Ac, and H3 relative to unmodified H4 across the 20-kb right terminal region of chromosome 6 (Chr6). (Middle) Diagram of coding genes found at these regions, according to assembly 22. (Bottom) Diagram depicting statistically significantly enriched (red) or depleted (blue) domains for each histone modification. (B) (Top) Fold enrichment (log2) of H3K4me3, H3K9Ac, H4K16Ac, and H3 relative to unmodified H4 at the rDNA locus and flanking regions (ChrR). (Middle) Diagram of coding genes (white) and ncRNAs (gray) found at this region, according to assembly 22. (Bottom) Diagram depicting statistically significantly enriched (red) or depleted (blue) domains for each histone modification. (C) Average profiles of histone modifications at MRSs and at upstream and downstream sequences. The gray arrow indicates the location of the FGR gene. For each histone modification, the fold enrichment (log2) relative to unmodified H4 is shown. (D) (Left) Diagrams of the structure of the C. albicans LTR and non-LTR retrotransposons. (Right) Chromatin signature of LTR and non-LTR retrotransposons. Average profiles and heat maps of histone modification signatures across each sequence are shown. The relative fold enrichment (log2) levels for each histone modification normalized to unmodified histone H4 or for aligned reads of immunoprecipitated (IP) sample normalized to aligned reads of input (I) sample (for RNAPII ChIP-seq) are displayed. The blue-to-red color gradient indicates high to low levels of enrichment in the corresponding region. MIN (minimum), −1.5 log2; MAX (maximum), +1.5 log2.