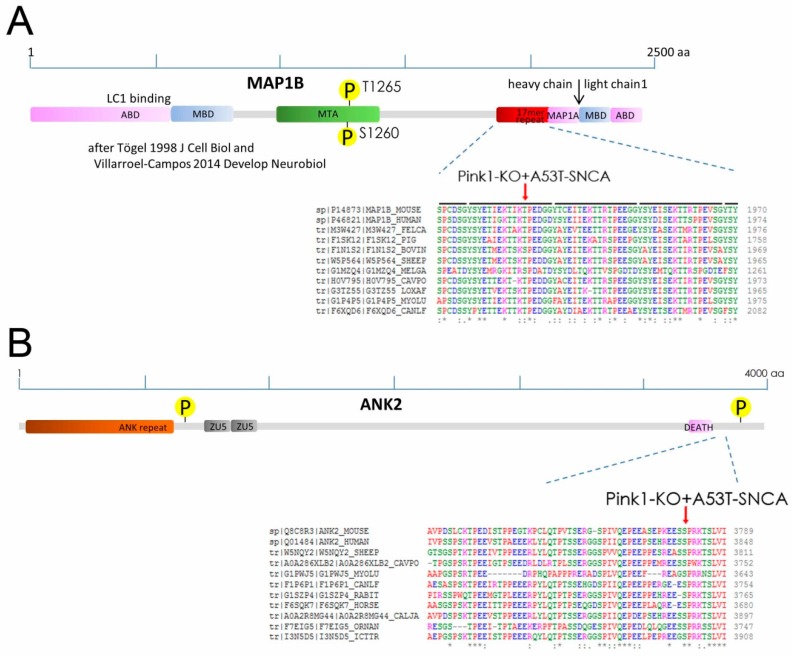
Figure 2.
(A) Scheme of MAP1B protein sequence, cleavage into heavy chain and light chain 1 (LC1), structural motifs with their interactions (LC1 binding in heavy chain, actin binding domain ABD, microtubule binding domain MBD, microtubule assembly helping site MTA, region that contains 12 repeats of a 17mer sequence, MAP1A interacting domain), and characterized GSK3beta-dependent phospho-sites at pS1260/pT1265, which have an established effect on cytoskeletal reorganization. Alignment of MAP1B protein sequences across species illustrates the conservation of the DM-modulated Thr/Ser site (red arrow) within the 17mer repeat (highlighted as black bars above sequence) of unknown function. The symbols * : . below the amino acid alignment highlight different phylogenetic conservation. (B) Scheme of ANK2 protein sequence, structural motifs (Ankyrin repeat, ZU5 domains, and DEATH domain, with characterized phosphorylation sites at S846 and S3850. Alignment of ANK2 protein sequences across species illustrates the conservation of the DM-modulated Ser site (red arrow) within the C-terminal target interaction domain. For a complete list of the residues with potential phosphorylation, see www.phosphosite.org.