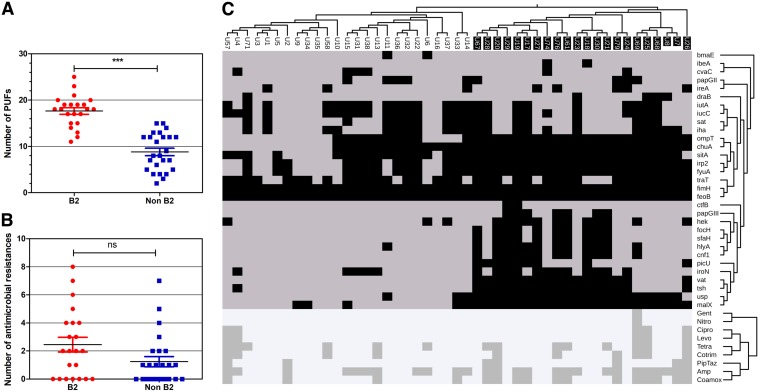
FIG 7.
Carriage of putative virulence factors (PUFs) but not antimicrobial resistance is associated with clade B2 E. coli urine isolates. (A) PUF scores differ between the B2 and non-B2 groups. ***, P < 0.0001, Mann-Whitney U test. (B) The antimicrobial resistance scores (number of different antimicrobials to which the strain is resistant) do not differ between the B2 and non-B2 groups. (C) The genome sequences of clinical urine E. coli isolates were screened for the presence of 31 previously described PUFs (y axis labels) using BLASTN analysis. The presence (black squares) or absence (gray squares) is shown for each PUF in relation to each isolate. Two-dimensional hierarchical clustering shows the PUF cooccurrence by strain (upper y axis dendrogram) and the PUF association with the phylogeny (x axis dendrogram). Clade B2 strains are indicated by white names on a black background (x axis labels). The lower diagram shows the hierarchical clustering of resistance (dark gray squares) and sensitivity (pale gray squares) to nine different antimicrobials (lower y axis dendrogram) by strain phylogeny. Abbreviations: Gent, gentamicin; Nitro, nitrofurantoin; Cipro, ciprofloxacin; Levo, levofloxacin; Tetra, tetracycline; Cotrim, co-trimoxazole; PipTaz, piperacillin-tazobactam; Amp, ampicillin; Coamox, amoxicillin-clavulanic acid.