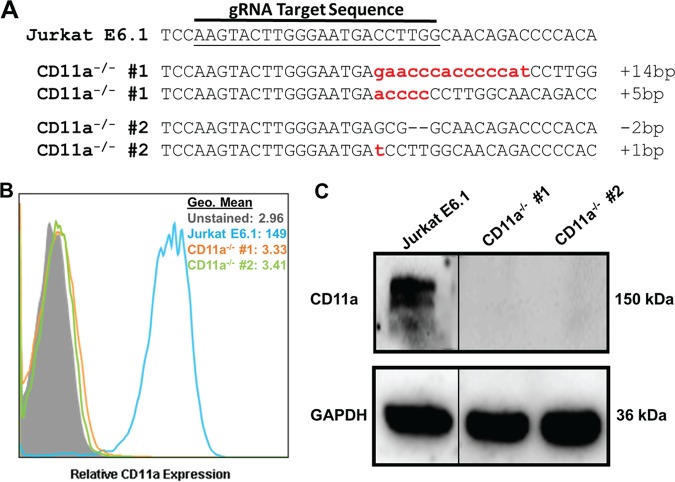
FIG 2.
Generation and validation of CD11a knockout in Jurkat cells. (A) DNA from the independent clones with mutations in CD11a was isolated, PCR amplified, cloned into a TOPO vector, and sequenced. Sequences were aligned to wild-type Jurkat E6.1 CD11a DNA. Any alteration in the DNA sequence was considered gene editing. Each dash indicates a single nucleotide deletion; lowercase letters indicate nucleotide insertions. The number of nucleotides inserted or deleted is indicated at the end of each sequence. (B) Flow cytometric analysis of Jurkat E6.1 cells and both Jurkat CD11a−/− clones for surface expression of CD11a. Cell populations are indicated according to the legend on the figure. (C) Western blot analysis to confirm deletion of CD11a and the GAPDH loading control. Gels were spliced for labeling purposes.