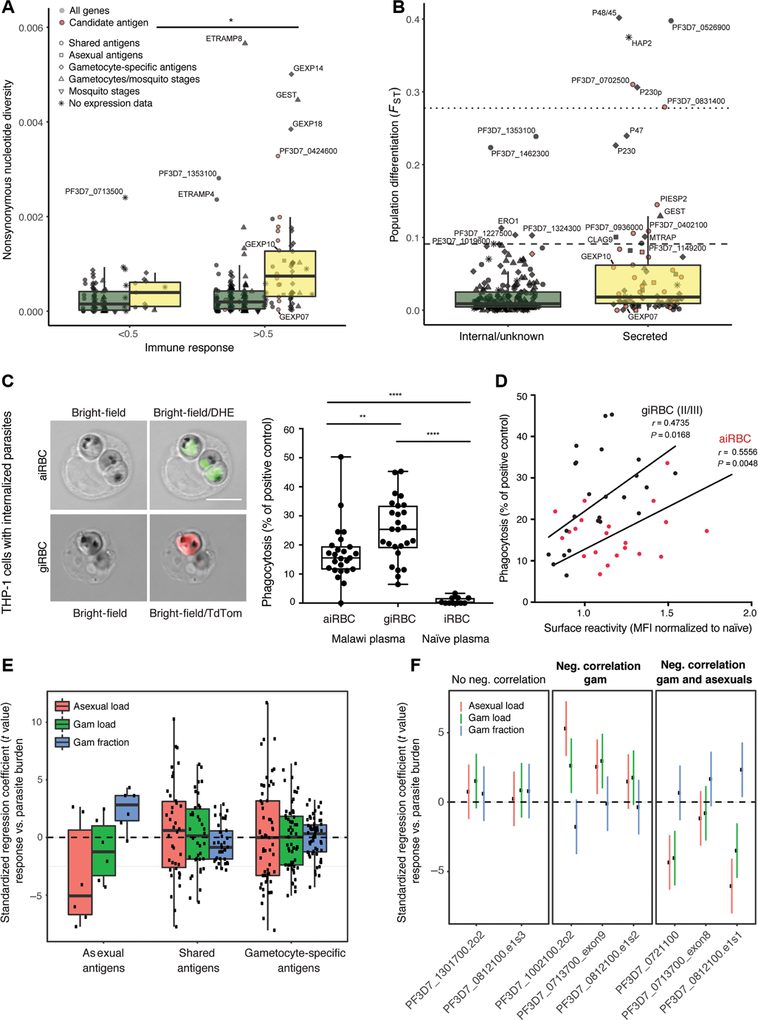
Fig. 6. Candidate gametocyte surface antigens elicit responses correlated with reduced gametocyte burden and a subset show minimal genetic diversity.
(A) Nonsynonymous nucleotide diversity for all antigens present on the protein array, stratified by stage, localization, and level of immune response (Mann-Whitney U test, P < 0.05). Genome data are from a set of parasite samples in Senegal. (B) Population differentiation between Senegal and Malawi parasite samples for secreted and internal proteins (FST at nonsynonymous sites; Mann-Whitney U test, P = 2.0 × 10−5). The dotted and dashed lines mark the 99th and 95th percentile of genome-wide nonsynonymous FST values. (C) Left: Internalized aiRBCs and giRBCs upon phagocytosis by THP-1 cells. aiRBCs are stained with the nuclear dye dihydroethidium (DHE) and giRBCs show TdTomato (TdTom) reporter fluorescence. Right: Phagocytosis index of Malawi plasma samples relative to positive control (rabbit antihuman RBC) and naïve U.S. serum. (D) iRBC phagocytosis versus surface recognition. (E) Associations were estimated between gametocyte fraction (gametocytes/total parasites), gametocyte and asexual parasite load, and secreted antigen fragment responses by peptide array [after normalization to in vitro transcription-translation (IVTT) controls and quantile normalization as in Fig. 1] across three malaria-exposed populations. Median standardized regression coefficient, −0.86; Wilcoxon test, P = 0.087. (F) Regression coefficients (and 95% confidence intervals) between individual protein fragments of the prioritized candidate antigens and parasite parameters (gametocyte fraction, gametocyte load, or asexual parasite load). Fragments are stratified by their correlation with parasite parameters. *P < 0.05, **P < 0.01, and ****P < 0.0001.