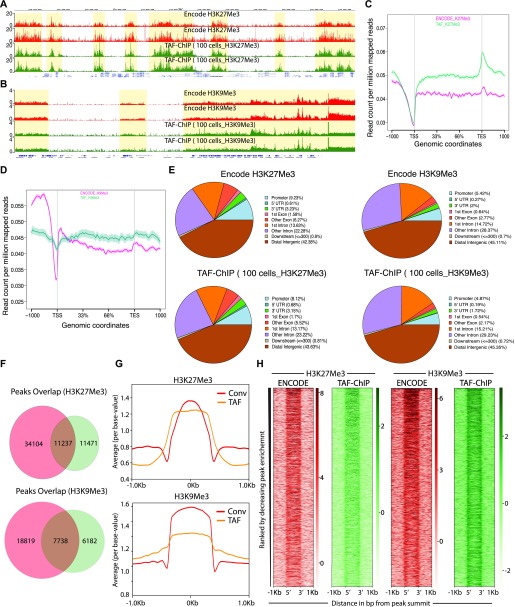
Figure 3. TAF-ChIP results from 100 K562 cells are comparable with conventional Encode ChIP-Seq datasets.
(A, B) Genome browser track example of H3K27Me3 and H3K9Me3 (A and B, respectively) ChIP performed in 100 FACS sorted K562 cells with TAF-ChIP approach and corresponding K562 conventional ChIP-Seq datasets from the ENCODE project in duplicates, as indicated in the labels. The label below the tracks shows the gene model and the y-axis represents normalized read density in reads per million. The enriched regions are highlighted with shaded box. (C, D) Metagene profiles of H3K27Me3 and H3K9Me3 (C and D, respectively) with standard error to the mean for all the genes, −1,000 bp upstream of TSS and +1,000 bp downstream of transcription end sites (TES). Read counts per million of mapped reads is shown on the y-axis, whereas the x-axis depicts genomic coordinates. (E) Genomic distribution of annotated peaks obtained from the ENCODE datasets and TAF-ChIP (100 K562 cells), for indicated histone marks. Note the majority of H3K27Me3 and H3K9Me3 peaks are at the intergenic regions, consistent with the expectation. (F) Overlap between the peaks identified from the ENCODE and TAF-ChIP datasets, for the indicated histone modifications (see the Materials and Methods section for further details). (G) Average profile of TAF-ChIP and corresponding ENCODE ChIP-Seq centered at the peaks for the indicated histone modifications. The y-axis depicts average per base value into the peaks, whereas x-axis depicts genomic coordinates centered at the peaks. (H) Distributions of reads at gene locations of indicated histone modifications from ENCODE ChIP-Seq and TAF-ChIP method, centered at the peaks (−1 kb to +1 kb). Rows indicate all the peaks and are sorted by decreasing affinities in the ENCODE ChIP-Seq datasets. The color labels to the right indicate the level of enrichment.