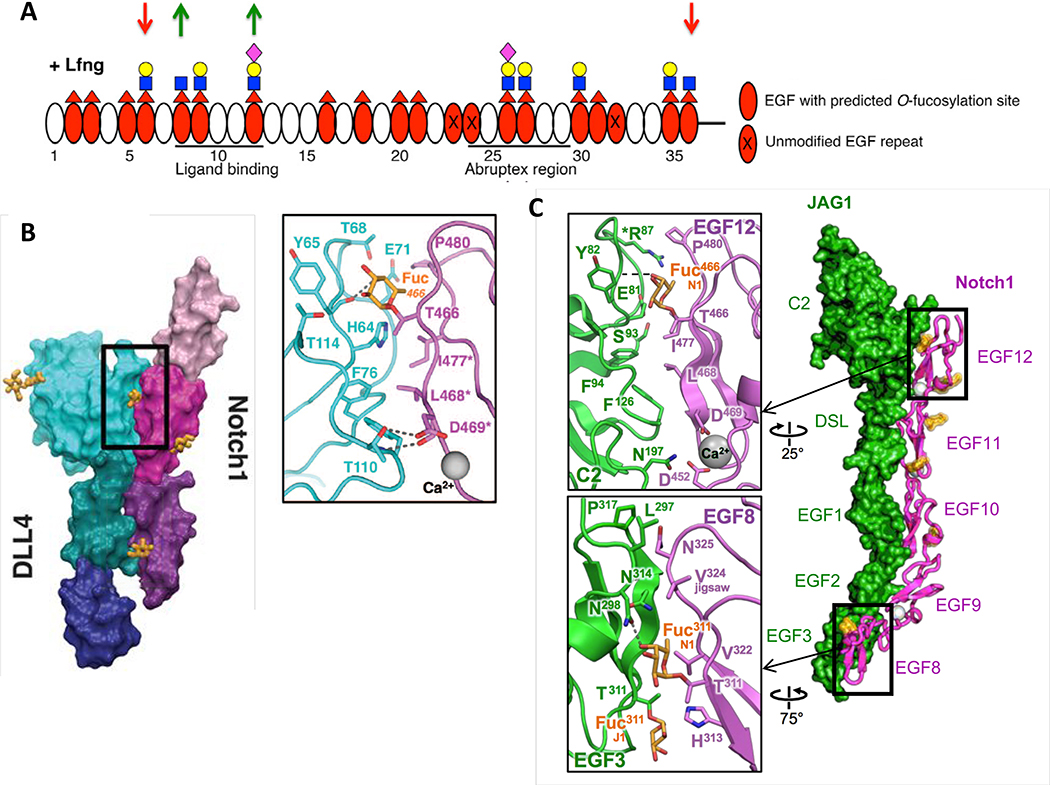
Figure 2. O-Fucose on EGF8 and EGF12 of NOTCH1 is in direct contact with ligands.
A. Domain map of mouse NOTCH1 EGF1–36 showing which EGF repeats are modified by O-fucose and elongated by LFNG (modified from [26]). MFNG elongates similarly, but RFNG only modifies O-fucose on EGF8, 12 and 26. Note that Fringe enzymes were overexpressed in a Fringe-deficient background in these studies. Down red arrows indicate sites where Fringe modification inhibits JAG1-NOTCH1 activation. Green up arrows indicate sites where Fringe modification enhances DLL1-NOTCH1 activation. Fucose, red triangle; GlcNAc, blue square; Galactose, yellow circle; Sialic Acid, purple diamond. B. Co-crystal structure of NOTCH1 EGF11–13 (shades of magenta/purple) and DLL4 (N-terminus to EGF3, shades of blue/green) (modified from [52]). Inset shows direct interaction between O-fucose on NOTCH1 EGF12 with residues in DLL4. C. Co-crystal structure of NOTCH1 EGF8–12 and JAG1 N-EGF3 (modified from [53]). Inset shows direct contacts between O-fucose on EGF8 and EGF12 and residues in JAG1. Note that the structures in B and C were obtained after directed evolution of the ligands toward stronger affinities.