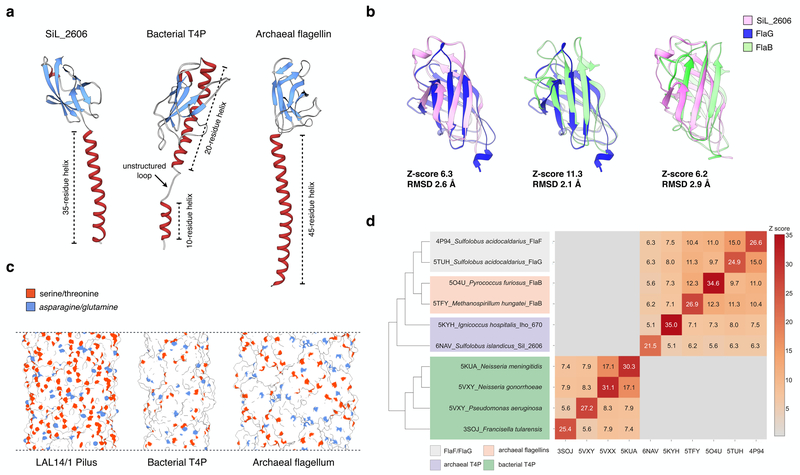
Figure 5. Comparison of LAL14/1 pilus with bacterial type IV pilus and archaeal flagellar filament.
a, ribbon display of a single subunit of Lal14 pilin, type IV pilin of N. gonorrhoeae (PDB 5VXX), and archaeal flagellin of M. hungatei (PDB 5TFY).
b, Structural alignment of the globular β-sandwich domains: LAL14/1 SiL_2606, FlaG (PDB 5TUH) and archaeal flagellin FlaB (PDB 5TFY). Z-scores are calculated by the DALI server.
c, Surface display of the filaments composed of pilins/flagellins shown in (a). The side chain oxygen atoms of serines and threonines are shown in orange, and the side chain nitrogen atoms of asparagines and glutamines are shown in blue.
d, All-against-all comparison of the bacterial and archaeal structures having N-termal T4P domains. The matrix (right) and cluster dendrogram (left) are based on the pairwise Z-score comparisons calculated using DALI. The color scale indicates the corresponding Z-scores. Based on their functions and origins, the proteins are partitioned into four groups, which are indicated with different colors.