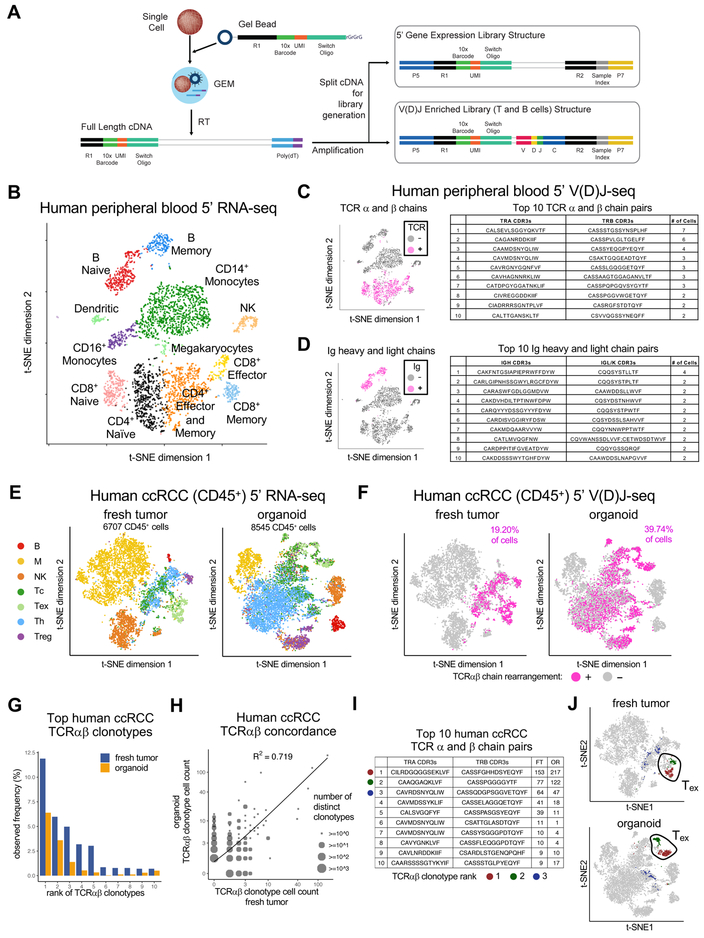
Figure 5. Droplet-based tandem single cell 5’ V(D)J and 5’ RNA-seq of immune cells.
(A) Chromium single cell Immune Profiling Solution. Single cell 5’ GEX and enrichment libraries can be generated from the same sample.
(B) t-SNE plot of 5’ scRNA-seq human healthy donor PBMCs.
(C, D) (Left) t-SNE plot of (B) with PBMCs having rearranged TCR (C) or Ig (D) clonotypes by single cell 5’ V(D)J-seq and T cell enrichment assay (magenta). (Right) Top 10 paired TCR (C) or Ig (D) clonotypes from T or B cells, respectively.
(E) t-SNE plots of 5’ RNA-seq of human ccRCC CD45+ FACS-sorted cells from fresh tumor (FT, left) or day 7 PDO (right).
(F) t-SNE plots of single cell 5’ V(D)J-seq of human ccRCC CD45+ cells from FT (left) vs. day 7 PDO (right). Cells with detected TCR clonotypes by 5’ V(D)J T cell enrichment assay from (E) are colored in magenta and correspond to T cell identity by 5’ scRNA-seq in (E).
(G) Observed frequency of top 10 TCR clonotypes in FT vs. PDO from (E) (ranked by order in FT).
(H) Scatter plot of cell counts between matching FT and PDO clonotypes from (F) (log scale). Circles with larger sizes indicate multiple overlapping data points, i.e., distinct clonotypes having same frequencies in fresh tumor and organoid. Concordance (R2 = 0.719) between FT and PDO is significant (p < 0.01, permutation test).
(I) Paired TCRαβ chain sequences and exact cell counts of the number of individual TILs expressing each unique TCR clonotype in F and G. The top 3 clonotypes are denoted by dark red, green and blue dots.
(J) t-SNE plots denoting the top 2 clonotypes in FT and PDO localize to exhausted T cells identified by 5’ RNA-seq in (E).
See also Figures S1–S5, Mendeley Figures 8–9, Table S3 and STAR Methods.