Figure 3.
Reproducible Assessments of Immune Composition across Independent Analyses
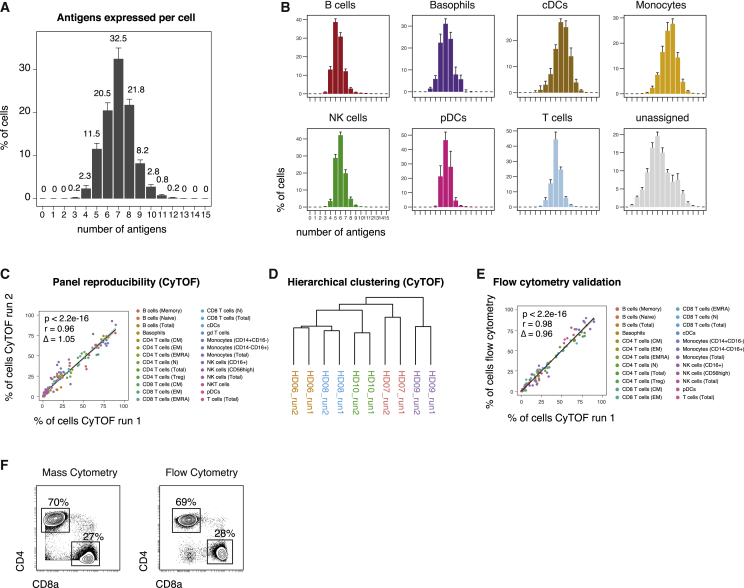
PBMCs from healthy donors (n = 5) were analyzed in two research centers. Immune cell populations were identified through serial gating as before (see Figure 2).
(A) Median number of positive antigens per cell, based on manually determined cutoffs (see Figure S2A). Numbers indicate median frequency of total pre-gated cells. Error bars represent SEM.
(B) Median number of positive antigens per cell as in (A), stratified by immune cell lineage.
(C) Different PBMC aliquots of the same donors (n = 5) were stained and acquired by mass cytometry in two different research institutes. Frequencies of immune lineages were determined through serial gating. Linear regression line is shown in black with the 95% confidence intervals (CIs, shaded). Coefficients, p values, and slope Δ were calculated based on data from all donors.
(D) Hierarchical clustering of samples from two independent mass cytometry runs based on frequencies as in (C).
(E) PBMCs aliquots of the same donors as in (C) were stained and acquired by flow cytometry, employing four separate staining reactions. Frequencies of immune lineages were determined through serial gating and plotted against the frequencies determined from mass cytometry as in (C). Linear regression line is shown in black with the 95% CIs (shaded). Coefficients, p values, and slope Δ were calculated based on data from all five donors.
(F) Exemplary biaxial plots and frequencies of CD4+ and CD8+ T cell subsets within one donor (HD08), as determined by mass cytometry (left) and flow cytometry (right).