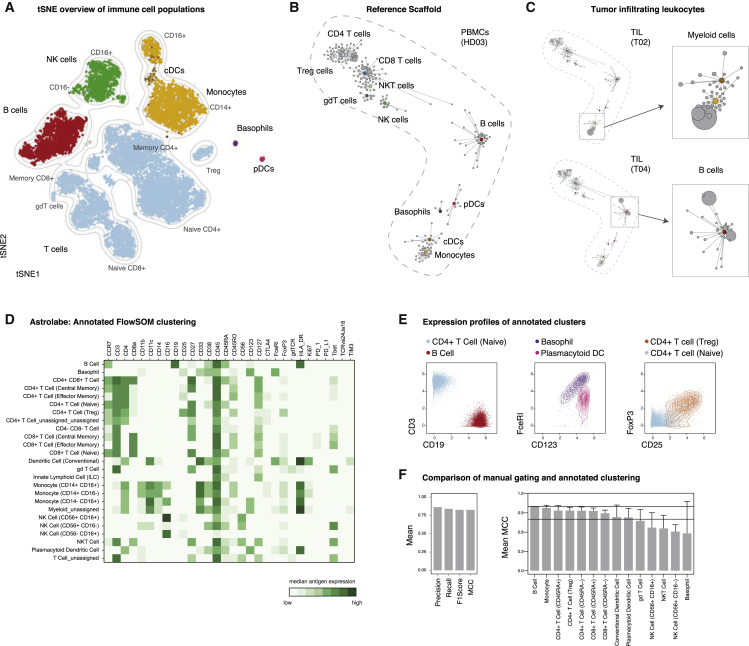
Figure 4.
Automated Data Visualization and Population Identification
PBMCs from healthy subjects (n = 5) and tumor biopsies from cancer patients (n = 5) were analyzed by mass cytometry using the reference panel (see Table S1).
(A) Data from all healthy donors was randomly subsampled to 20,000 cells and subjected to tSNE dimensionality reduction. Cells are colored by their immune cell lineage assignment from manual gating. Grey indicates cells unassigned by manual gating.
(B) A reference scaffold map of PBMC data was created using manually gated landmarks (colored) and all antigens for the clustering analysis. Inter-cluster connections were used to create the graph but are not depicted here. Shown is one representative sample (HD03).
(C) Pre-gated, CD45+ cells from tumor samples were mapped onto the reference scaffold. Maps from two patients are shown (left). Enlarged examples of modulated immune cell populations are pointed out (right).
(D) PBMC data as above were clustered and automatically annotated using the Astrolabe platform. Shown are median expression levels of all antigens for all clusters.
(E) Exemplary expression profiles of immune cell populations as determined by Astrolabe (HD06).
(F) Mean precision, recall, F1score, and Matthews correlation coefficient (MCC; see Method Details) between manual lineage assignments and FlowSOM-based clustering for all donors and populations (left). Mean MCC for all donors stratified by population (right). Two horizontal lines indicate MCC = 1 (maximum agreement) and MCC = 0.8, respectively. Error bars represent SEM.