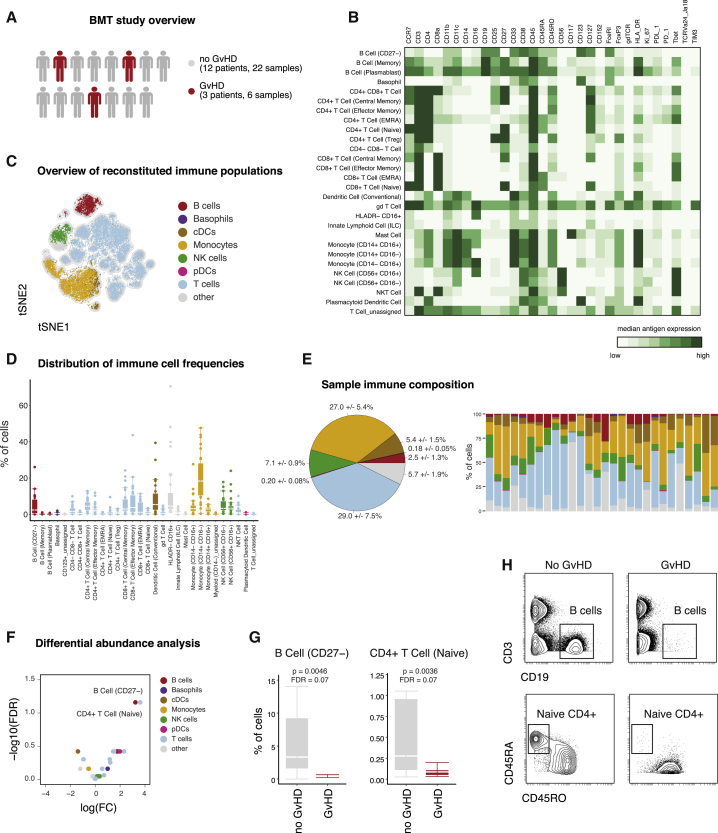
Figure 5.
Identification of Disease-Associated Immune Signatures Following Bone Marrow Transplantation
(A) Following tumor therapy, patients (n = 15; Table S2) underwent bone marrow transplantation. Peripheral blood samples were collected and subsequently stained with the described reference panel and analyzed by mass cytometry.
(B) Data were uploaded to the Astrolabe platform, clustered, and automatically annotated. Exemplary heatmap of one patient depicting the median protein expression levels across all populations identified through clustering.
(C) The 20,000 randomly subsampled cells of one patient were subjected to tSNE dimensionality reduction. Color-assignments represent different immune lineages as identified through annotated clustering.
(D) Clustering-derived frequencies of immune populations for all samples in this study (n = 28). Boxplots depict the interquartile range (IQR) with a horizontal line representing the median. Whiskers extend to the farthest data point within a maximum of 1.5× IQR. Points represent individual samples.
(E) Frequencies of immune cell subpopulations were combined into frequencies for major immune cell lineages and color-coded as in (C). Pie chart depicting the median frequencies ± SEM of all major immune lineages across all samples (left). Immune composition for all analyzed samples (n = 28; right). (F) FDR and fold change (FC) of immune cell frequencies in patients with or without GvHD.
(G) Comparison of differentially abundant immune cell frequencies in patients with or without GvHD. Boxplots depict the IQR with a horizontal line representing the median.
(H) Confirmation of reduced abundance of B cells (top) and naive CD4+ T cells (bottom) in an exemplary patient with (right) and without (left) GvHD. Examples of B cells were pre-gated on single, live, CD45+ cells. Examples of naive CD4+ T cells were pre-gated on single, live, CD4+ T cells.