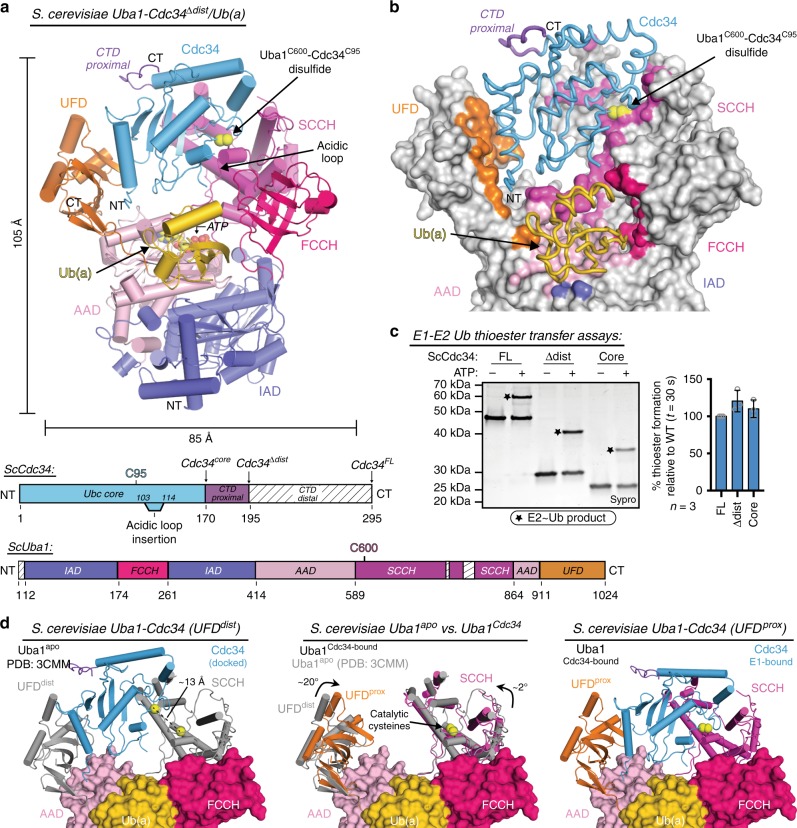
Fig. 1.
Overall architecture of a ScUba1-Cdc34Δdist/Ub(a) complex. a Top, overall structure of S. cerevisiae Uba1 in complex with truncated S. cerevisiae Cdc34, Cdc34Δdist (blue), and ubiquitin adenylate, Ub(a) (gold), with Uba1 domains colored and labeled. Catalytic cysteines are indicated by yellow spheres. Bottom, schematic representations of the ScUba1 and ScCdc34 constructs with domains colored as above. b Uba1-Cdc34 structure with Uba1 surface representation in gray with only contacting residues colored. Cdc34Δdist and Ub(a) are represented as worms. c E1-E2 thioester transfer assays for Cdc34 variants with boundaries as indicated in schematic, and with normalized quantification of E2 thioester formed represented as mean ± SD with three independent replicates shown as gray circles, right. Stars indicate the E2~Ub thioester product. Source data are provided as a Source Data file. d Left, Cdc34E1-bound modeled onto Uba1apo UFD (gray) in open conformation (PDB:3CMM); distance between catalytic cysteines is indicated by a dashed line. middle, ScUba1Cdc34-bound/Ub(a) as in a with ScUba1apo/Ub(a) superimposed in gray. Arrows indicate rotation of the UFD and SCCH when bound to Cdc34. Right, ScUba1-Cdc34Δdist/Ub(a) complex structure colored as in a