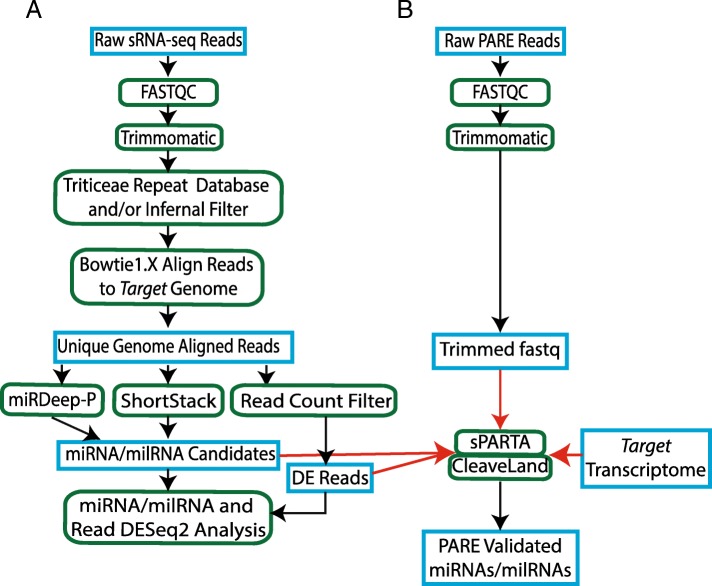
Fig. 1.
Small RNA sequencing and PARE sequencing analysis pipelines. (A) Small RNA-Seq Illumina reads were trimmed, filtered, and run through the two plant miRNA identification programs miRDeep-P and ShortStack to identify miRNA/milRNA candidates and DE reads. (B) Sequencing reads from the PARE libraries were trimmed and filtered and analyzed with the sPARTA version 1.21 [31] and CleaveLand (version 4.4) [32]. Additional input data was provided from the barley or Blumeria transcriptome and miRNA/milRNA candidates plus DE reads developed from the sRNA sequencing pipeline. Input or output files are highlighted with blue boxes, programs or processes are highlighted with green ovals, and PARE program inputs are highlighted with red arrows