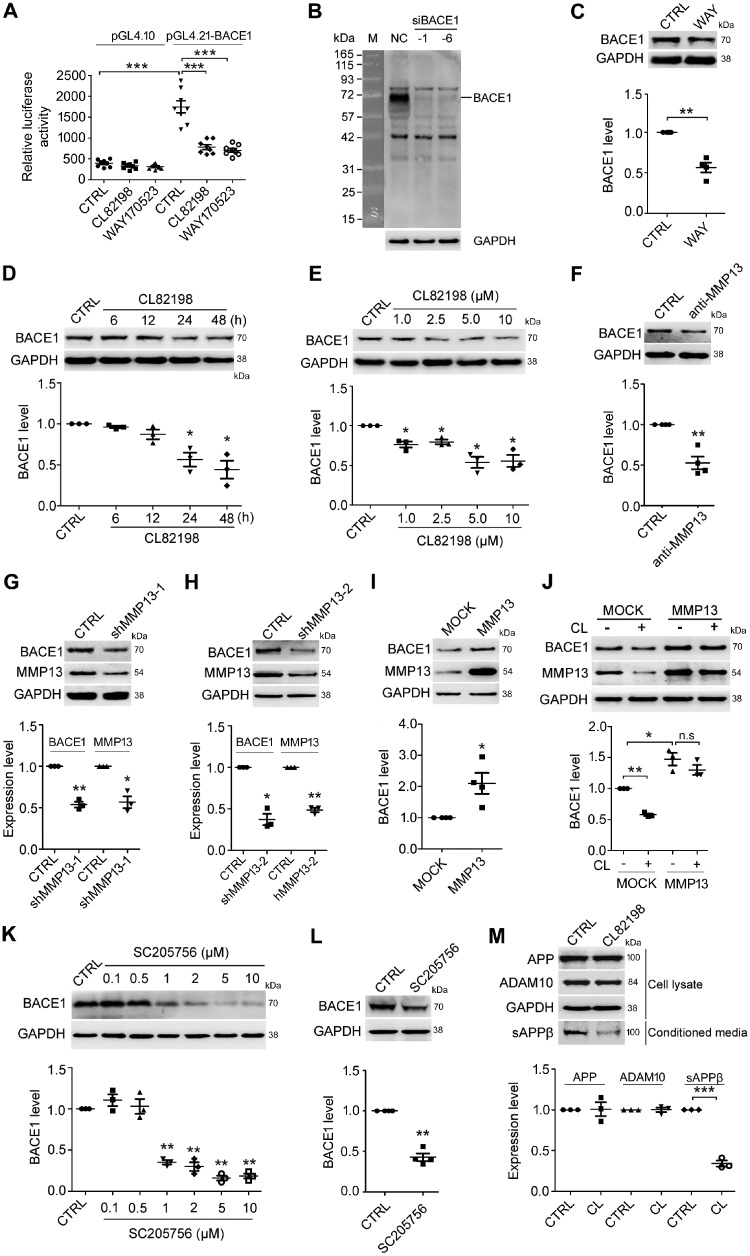
Figure 1.
CL82198 inhibits BACE1 protein expression through MMP13. (A) HEK293 cells were transfected with the luciferase reporter plasmids pGL4.21-BACE1 and pGL4.10 (negative control) in the absence or presence of 5 μM CL82198 or 10 nM WAY170523 for 48 h. Luciferase assays were performed with a GloMax 96 microplate luminometer. Firefly luciferase activity was normalized to that of the plasmid pGL4.10. (B) BACE1 was knocked down with BACE1 siRNA (siBACE-1 and -6) or control siRNA (NC) in HEK293 cells for 48 h. A dramatic decrease in the amount of BACE1 protein is shown at ∼70 kD. M = protein marker. (C) HEK293 cells were treated with WAY170523 (WAY, 10 nM) for 48 h, while control cells were treated with DMSO (CTRL). (D) SH-SY5Y cells were treated with 5 μM CL82198 for 6, 12, 24 and 48 h. (E) SH-SY5Y cells were treated with 1, 2.5, 5 and 10 μM of CL82198 for 48 h. (F) SH-SY5Y cells were treated with an MMP13 neutralizing antibody (anti-MMP13, 1:500) or control rabbit IgG antibody (CTRL) for 48 h. (G–I) Mmp13 was knocked down with shRNA-1 (shMMP13-1, G) or shRNA-2 (shMMP13-2, H) in HT22 cells for 72 h or was overexpressed with an MMP13 vector in HEK293 cells for 48 h (I). CTRL = control shRNA; MOCK = control vector. (J) HEK293 cells were transfected with either the control vector (MOCK) or MMP13 vector for 48 h in the absence or presence of 5 μM CL82198 (CL). Cell lysates were subjected to western blotting analysis. Representative western blots for BACE1 are shown on the top, and quantifications are shown below. (K) Primary cultured cortical neurons were treated with 0.1, 0.5, 1, 2, 5 and 10 μM SC205756 for 48 h. (L) HEK293 cells were treated with 1 μM SC205756 for 48 h. (M) HEK-APP cells were treated with 5 µM CL82198 (CL) for 48 h. Cell lysates were prepared and subjected to western blotting analysis for APP and ADAM10. sAPPβ was analysed in conditioned media using an sAPPβ antibody. All values were normalized to CTRL or MOCK (1.0) within each experiment. The error bars are the SEM. n.s. = no significant difference; *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA, n = 3 or 4).