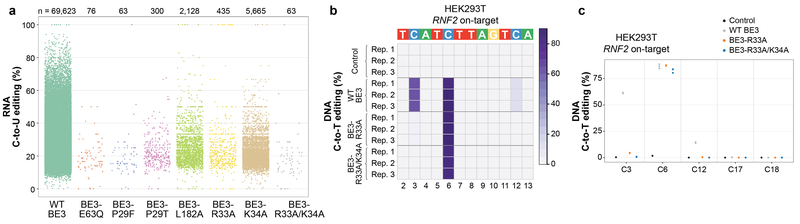
Extended Data Figure 6. Additional data showing SECURE-BE3 variants induce substantially reduced numbers of RNA edits but possess comparable and more precise DNA editing activities in HEK293T.
(a) Initial screen of the transcriptome-wide RNA editing activities of six BE3 variants harboring various APOBEC1 mutations and expressed at high levels in HEK293T cells (sorting cells with top 5% of GFP signal). Jitter plots of single replicate RNA-seq experiments showing RNA cytosines modified by expression of wild-type (WT) BE3, BE3-E63Q (APOBEC1 catalytic site mutant), BE3-P29F, BE3-P29T, BE3-L182A, BE3-R33A, BE3-K34A, and BE3-R33A/K34A variants. Y-axis represents the efficiencies of C-to-U editing. n = total number of modified cytosines observed in each sample. (b) Heat map of on-target DNA base editing efficiencies of WT BE3, BE3-R33A, BE3-R33A/K34A, and nCas9-UGI-NLS (Control) in HEK293T cells with the RNF2 gRNA (cells from same experiment as presented in Fig. 2a). Bases within the editing window of the on-target spacer sequence are numbered as previously described. Note the inclusion of C12, which is inefficiently edited by WT BE3 in these samples but not edited by the SECURE-BE3 variants, even in the higher expression context. (c) Dot plot for HEK293T on-target data displayed in (b), expanded to include all cytosines across the spacer sequence.