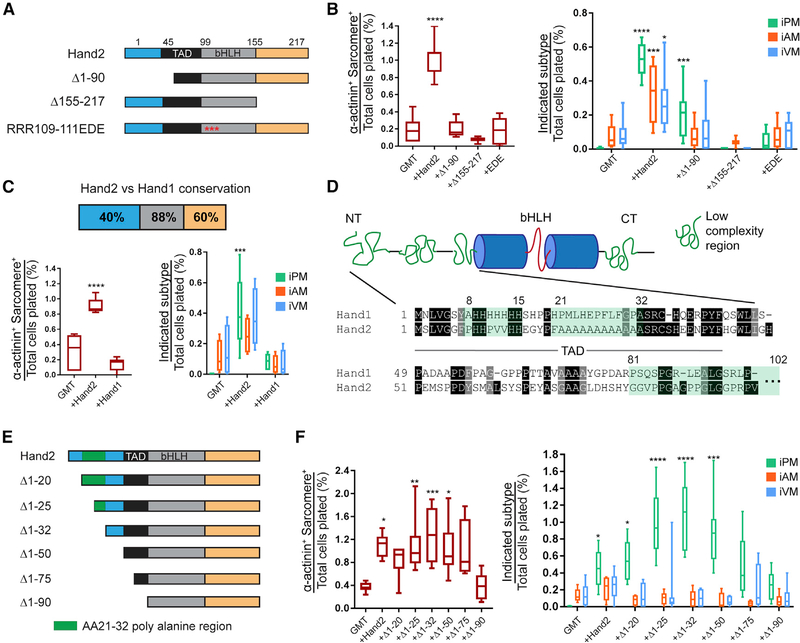
Figure 5. The Hand2 N Terminus Encodes a Cardiac Subtype Diversification (CSD) Domain.
(A) Schematic for the Hand2 mutants used in (B). Unstructured N and C termini are shown in blue and orange, respectively, whereas the known transactivation domain (TAD) is shown in black. Beta helix-loop-helix (bHLH) domain is shown in gray, and mutations in DNA-contacting basic residues in the RRR109–111EDE construct are depicted as red asterisks.
(B) Cardiac reprogramming with GMT alone or with the indicated Hand2 mutants was assessed for sarcomere organization (left) and subtype diversification (right) (means ± SD, n = 8). All samples were compared with GMT to determine statistical significance.
(C) Cardiac reprogramming was conducted with GMT plus Hand2 or Hand1, and iCLMs were assessed for sarcomere organization (left) and subtype diversity (right). The degree of primary amino acid conservation between Hand2 and Hand1 for each protein domain is indicated at the top (means ± SD, n = 6). All samples were compared with GMT to determine statistical significance.
(D) Cartoon diagram of Hand2 predicted secondary structure with low-complexity regions (LCRs) highlighted in green and bHLH region in blue cylinders and red line (top). The primary sequence of the N terminus is shown below with the indicated LCRs.
(E) Schematic for the Hand2 N-terminal mutants used for reprogramming in (F).
(F) Cardiac reprogramming was performed with GMT alone or combined with the indicated Hand2 N-terminal mutants. iCLMs were assessed for sarcomere organization (left) and subtype diversity (right) (means ± SD, n = 6). All samples were compared with GMT to determine statistical significance.
Statistical significance indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Non-significant comparisons indicated either by “ns” or no asterisk.