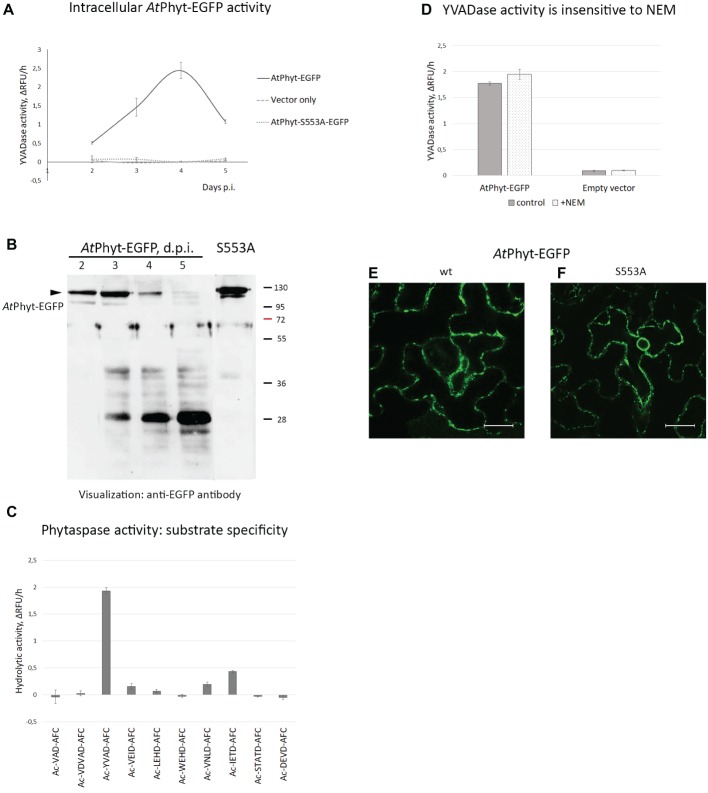
Figure 6.
Origin of phytaspase activity in AtPhyt-EGFP producing N. benthamiana leaves. (A) Determination of phytaspase activity in intracellular protein fractions obtained at various days p.i. from leaves producing AtPhyt-EGFP (bold line). Intracellular protein fractions from N. benthamiana leaves infiltrated with Agrobacterium cells carrying the empty vector (pCambia1300, dashed line) and total protein fractions from leaves producing the inactive AtPhyt-S553A-EGFP mutant (dotted line) served as controls. Fluorogenic peptide substrate Ac-YVAD-AFC was used at 30 μM to quantify the phytaspase activity. Relative rates of hydrolysis were determined as an increase of relative fluorescence units per hour (ΔRFU/h). Data represent the mean of three experiments ±SD. (B) Western blot analysis of AtPhyt-EGFP in total extracts from N. benthamiana leaves producing the AtPhyt-EGFP protein at various days p.i. The arrowhead points to position of full-length protein (~120 kDa). S553A, sample from leaves producing catalytically inactive AtPhytS553A-EGFP protein at 3 d.p.i. Monoclonal anti-EGFP antibody was used for protein detection. Positions of molecular weight markers are indicated on the right. (C) Cleavage specificity of the proteolytic activity under study corresponds to that of AtPhyt. Total protein extract obtained at day 4 p.i. was incubated with a panel of fluorogenic peptide substrates (30 μM). Ac-YVAD-AFC and Ac-IETD-AFC are the preferred AtPhyt substrates (Chichkova et al., 2018). Relative rates of hydrolysis were determined as in (A). Data represent the mean of three experiments ±SD. Specificity profiles with protein samples obtained at day 3 and 5 p.i. were similar to this one. (D) N-ethylmaleimide (2 mM), an inhibitor of VPE protease, does not interfere with hydrolysis of Ac-YVAD-AFC by total protein extracts obtained at day 4 p.i. from AtPhyt-EGFP producing (AtPhyt-EGFP) and non-producing (Empty vector) leaves. Relative rates of hydrolysis were determined as in (A). Data represent the mean of two experiments ±SD. Similar results were obtained with a 3 d.p.i. sample. (E,F) The AtPhyt-EGFP protein (wt, E) and its catalytically inactive mutant AtPhytS553A-EGFP (S553A, F) are produced with similar efficiency in N. benthamiana leaves. Images were obtained at day 2 post-infiltration. Bar, 20 μm.