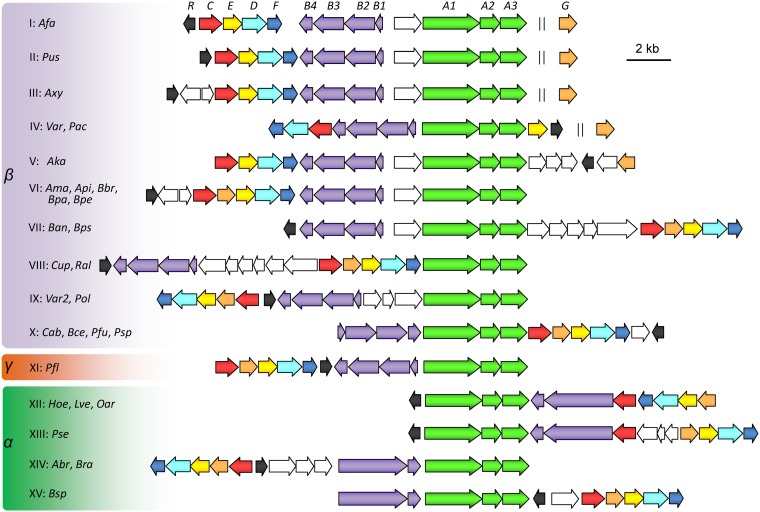
FIG 6.
Predicted PA catabolism gene clusters in bacterial genomes. α, β, and γ indicate the Alpha-, Beta-, and Gammaproteobacteria. I to XV, the 15 different types of PA catabolism loci. Abbreviations and representative strains are as follows: Afa, Alcaligenes faecalis JQ135; Pus, Pusillimonas sp. strain T2; Axy, Achromobacter xylosoxidans A8; Var, Variovorax paradoxus S110; Pac, Pseudacidovorax sp. strain RU35E; Aka, Advenella kashmirensis W13003; Ama, Achromobacter marplatensis B2; Api, Achromobacter piechaudii ATCC 43553; Bbr, Bordetella bronchiseptica RB50; Bpa, Bordetella parapertussis ATCC BAA-587; Bpe, Bordetella pertussis Tohama I; Ban, Bordetella ansorpii NCTC13364; Bps, Bordetella pseudohinzii HI4681; Cup, Cupriavidus necator N-1; Ral, Ralstonia pickettii DTP0602; Var2, Variovorax sp. JS1663; Pol, Polaromonas sp. strain OV174; Cab, Caballeronia arvi LMG 29317; Bce, Burkholderia cepacia JBK9; Pfu, Paraburkholderia fungorum NBRC 102489; Psp., Pandoraea sputorum DSM 21091; Pfl, Pseudomonas fluorescens C8; Hoe, Hoeflea sp. IMCC20628; Lve, Loktanella vestfoldensis SMR4r; Oar, Octadecabacter arcticus 238; Pse, Paracoccus seriniphilus DSM 14827; Abr, Afipia broomeae GAS525; Bra, Bradyrhizobium canariense GAS369; Bsp., Bradyrhizobium sp. strain C9. The detailed genomic accession numbers and the gene locus tags are listed in Table S1 in the supplemental material. Identities (percent) of amino acid sequences between Pic proteins of strain A. faecalis JQ135 and representative homologs are listed in Table S2.