Figure 3. Structure of Polη incorporating dCTP opposite the 3′G of the Pt(DACH)GpG adducts.
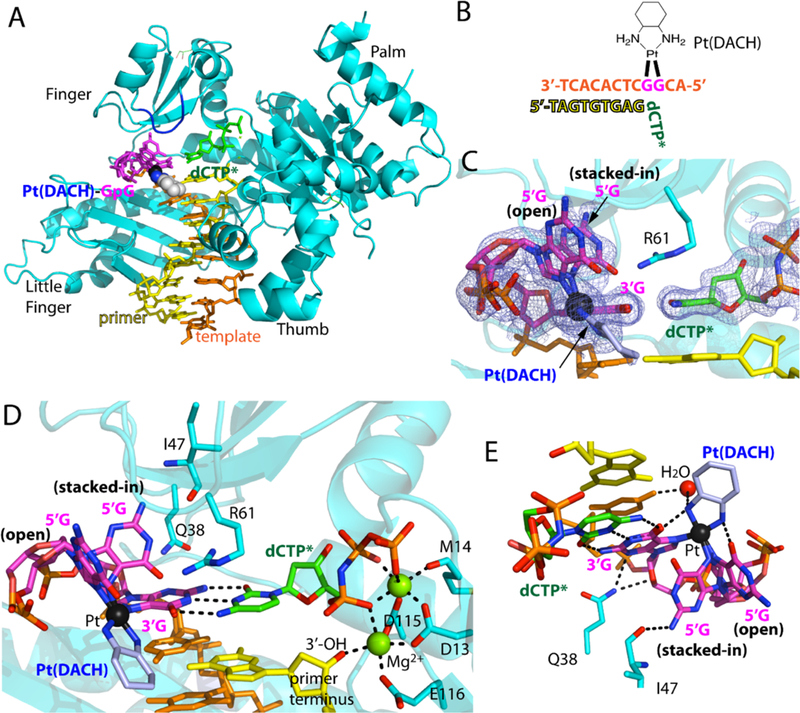
(A) Overall structure of Polη incorporating dCTP* opposite Pt(DACH)−3′G. The protein chain is colored in cyan, the template DNA that contains the two modified guanines (magenta) is colored in orange, the DNA primer in yellow, and the incoming nucleotide in green. The Pt(DACH) moiety is shown in spheres. The Val59-Trp64 loop near the Pt(DACH)GpG is shown in blue. (B) DNA sequences used for the crystallographic studies. (C) A 2Fo-Fc electron density map contoured at 1σ around the Pt(DACH):dCTP* base pair. Pt is shown in black sphere. The 5′G of the Pt(DACH)GpG exists as a mixture of an “open” and “stacked-in” conformations. (D) A close-up view of the active site of the Pt(DACH)−3′G-Polη complex. The catalytic triad (Asp13, Asp115, and Glu116) and two Mg2+ metal ions (binding and catalytic metals) are indicated. The incoming nonhydrolyzable dCTP* forms Watson-Crick base pair with the Pt(DACH)−3′G. The primer terminus dG is shown in yellow. Hydrogen bonds are indicated as dashed lines (E) Interaction of the Pt(DACH) moiety with the surrounding environment.
