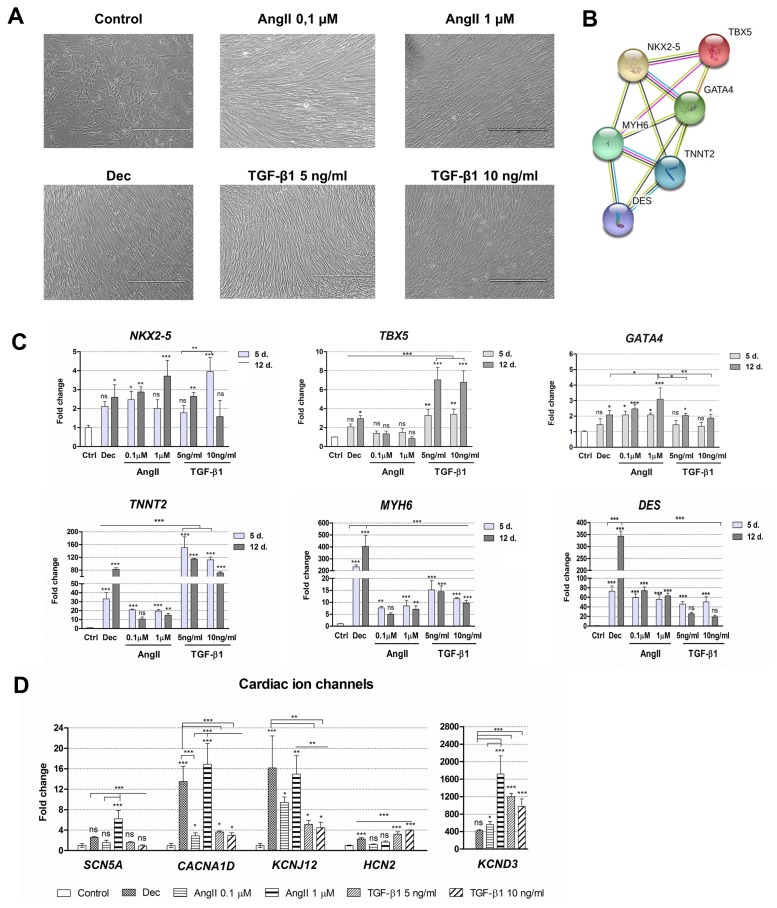
Fig. 2.
Alterations in morphology and gene expression patterns during decitabine, angiotensin II and TGF-β1 induced cardiac differentiation. (A) Assessment of the cell morphology 12 days after the cardiac differentiation initiation. Dec: decitabine, AngII: angiotensin II, TGF-β1: transforming growth factor-beta 1. Scale bar=400 μm. (B) The connections between proteins that are considered as cardiomyogenic differentiation markers. NKX2-5, TBX5 and GATA4 are the early transcription markers, initiating the expression of structural proteins of cardiomyocytes, such as MYH6 (α-myosin heavy chain), TNNT2 (cardiac troponin T) and DES (Desmin). The data was obtained from the STRING database. (C) The relative expression of the main cardiac : genes-markers, such as NKX2-5, TBX5, GATA4, MYH6, TNNT2 and DES at the days 5 (5 d.) and 12 (12 d.) of induced differentiation. Ctrl–non-differentiated control cells. (D) The relative expression of cardiac ion channels genes at the day 12 of differentiation: SCN5A (sodium voltage-gated channel α-subunit 5), CACNA1D (L-type calcium channel), KCNJ12 and KCND3 (voltage-gated potassium) and HCN2 (hyperpolarization-activated cyclic nucleotide-gated channel). The relative gene expression was determined by RT-qPCR, normalized to GAPDH and presented as a fold change over undifferentiated control. The data are presented as mean±SD (n=3), p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant.