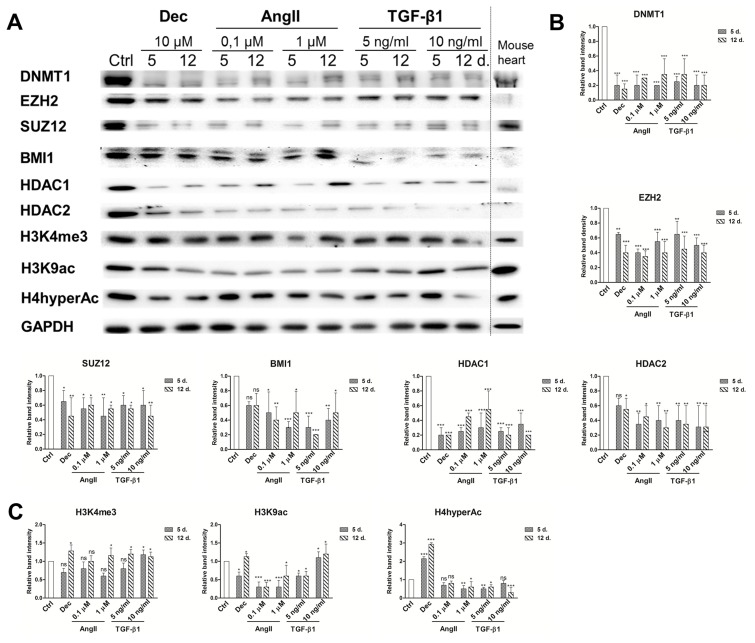
Fig. 5.
Epigenetic alterations during induced cardiomyogenic differentiation. (A) Total proteins were extracted from control cells (Ctrl) and cells, induced using angiotensin II and TGF-β1 at the days 5 and 12 of differentiation. Alterations in the expression levels of Polycomb repressive complex 2 (PRC2) proteins EZH2 and SUZ12, Polycomb repressive complex 1 (PRC1) component BMI1, DNA and histones modifying proteins DNMT1, HDAC1 and HDAC2 as well as in histone modifications, considered as the markers of active chromatin state (H3K4me3, H3K9Ac and H4hyperAc) are presented as determined by Western blot. Mouse heart lysate was used as a positive control. The blots represent one of three independent experiments showing similar results. (B) Relative band density of chromatin remodeling proteins and (C) histone modifications, as measured using ImageJ software (NIH, USA) and normalized to the GAPDH loading control. The data are presented as mean±SD (n=3), p≤0.05 (*), p≤0.01 (**), p≤0.001 (***), ns: non-significant.