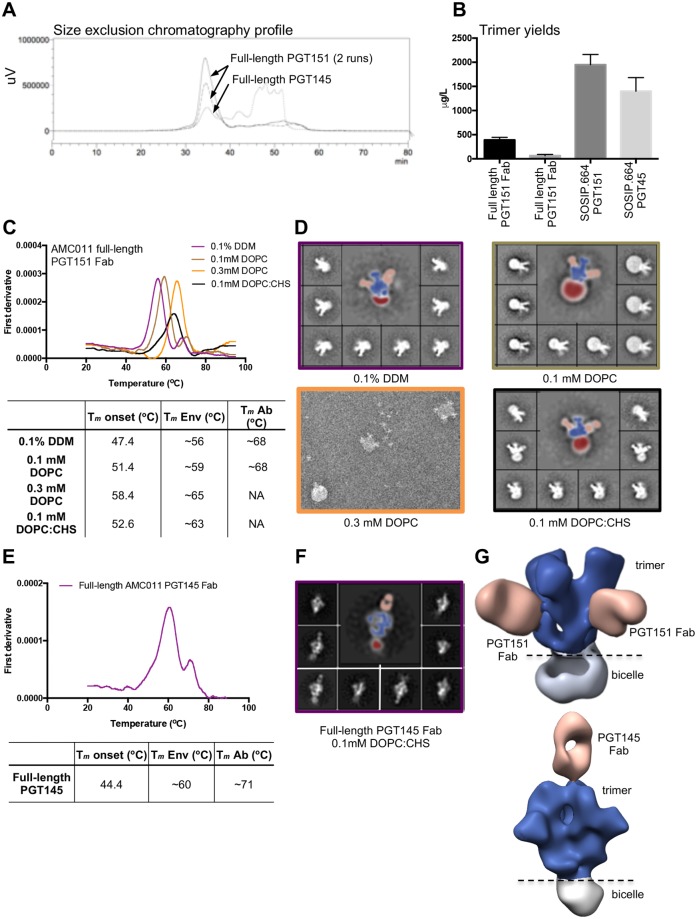
Fig 1. Purification of the full-length AMC011 trimer in complex with PGT151 or PGT145 Fab.
(A) Size exclusion chromatography profiles of full-length AMC011 Env bound to PGT151 Fab (two runs) or bound to PGT145 Fab (one run). (B) Average yields of full-length Env constructs bound to PGT151 Fab or PGT145 Fab and their SOSIP.664 counterparts. For comparison, soluble AMC011 SOSIP.664 trimers were purified with PGT151 and PGT145 affinity chromatography columns. The mean yields shown in the graph represent the averages of three to six purifications. (C) Thermostability of full-length AMC011 trimers in DDM or at different concentrations of DOPC and DOPC:CHS (1:1 molar ratio) after detergent removal. The unfolding pattern of the different proteins was assessed by plotting the first derivative of the curves acquired with nano-DSF. The Tm onset, the melting temperatures of the Env (Tm Env) and the Tm of the antibody (Tm Ab) are shown in the table. The averages Tm values derived from two to five different experiments are listed in the table. The raw data of one such experiment is depicted in the graph. NA: not applicable. (D) 2D class averages from negative-stain EM images are shown for the full-length AMC011 Env trimer bound to PGT151 Fab in DDM or at different concentrations of DOPC or DOPC:CHS (1:1 molar ratio). Colors in the thermostability graph match the 2D class average boxes. (E) Thermostability of full-length AMC011 Env trimer bound to PGT145 Fab in 0.1 mM DOPC:CHS (1:1 molar ratio). The Tm was calculated from one experiment. (F) Representative 2D class averages from full-length AMC011 Env trimer in 0.1 mM DOPC:CHS. (G) 3D models of full-length AM0C11 trimer bound to PGT151 or to PGT145, which were derived from the class-averages shown in (D) and (F). Coloring in (D), (F) and (G) is as follows: PGT151 and PGT145 in light orange, the bicelle in gray and the envelope trimer in dark blue.