Fig. 4. Nrf2-dependent changes of basal lung transcriptome by sulforaphane (SFN) pretreatment.
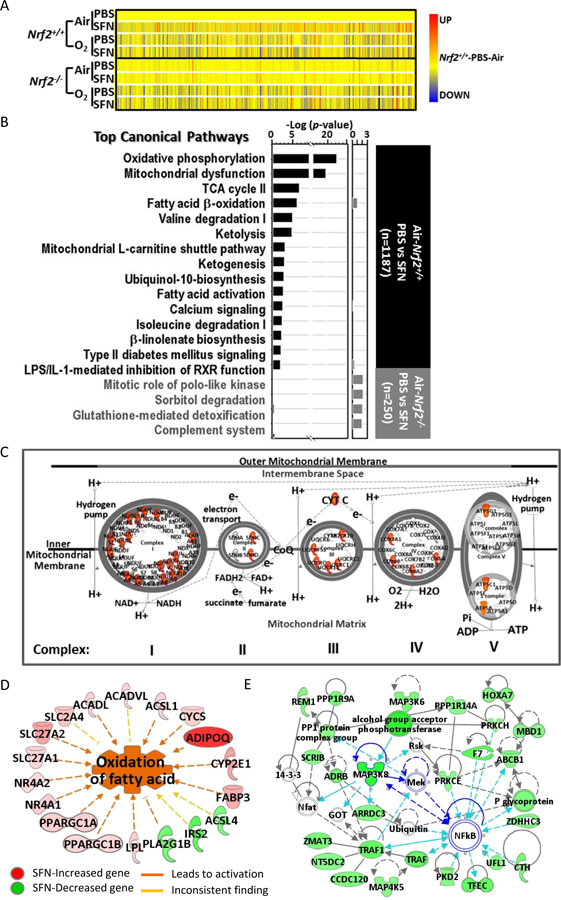
(A) Heat map from hierarchical clustering analysis depicts expression profiles of SFN-responded genes in Nrf2+/+ after 72 h hyperoxia (O2) exposure (n = 1,187, P < 0.01 with moderated t-test). Heat map for the same genes in Nrf2−/− mice were shown for comparison. Color bar indicates average expression intensity (n = 3/group) normalized to Nrf2+/+-PBS-Air group. (B) Top canonical pathways of lung genes significantly altered by SFN in air-exposed Nrf2+/+ (black bars) and in air-exposed Nrf2−/− mice (grey bars). (C) Mitochondrial oxidative phosphorylation complex is illustrated with genes (in red) that were induced by SFN treatment in Nrf2+/+ mice exposed to normoxia (room air). (D) Top diseases and bio-functions of SFN-responsive lung genes in Nrf2+/+ mice included energy metabolism such as fatty acid beta-oxidation. (E) Lung genes significantly reduced by SFN in Nrf2+/+ mice were involved in the network of organismal injury and abnormality (scores 32–41), in which key molecules such as TNF receptor associated factor 1 (Traf1) and multiple mitogen-activated protein kinase (MAPK) cascade enzymes (e.g., Map3k8) were predicted to play central roles with NF-κB. Analysis was done by Ingenuity Pathway Analysis software.
