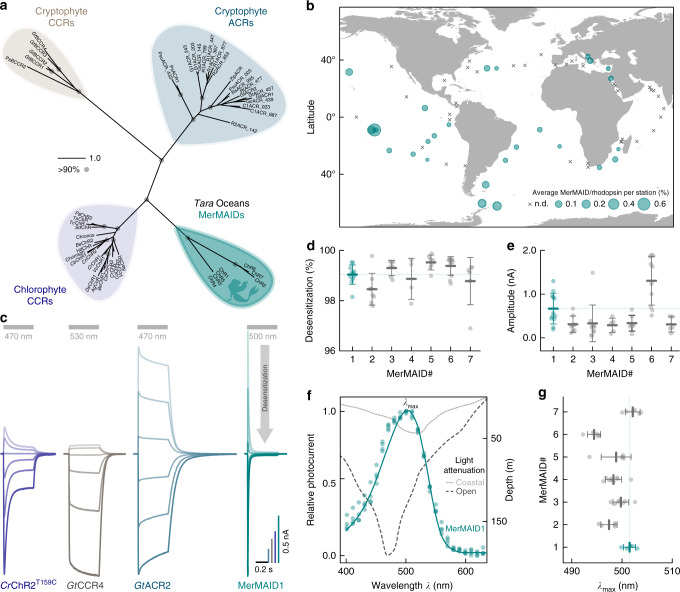
Fig. 1.
Discovery and electrophysiological features of MerMAID.s a Unrooted phylogenetic tree of the channelrhodopsin superfamily, with gray circles representing bootstrap values >90%. Scale bar indicates the average number of amino acid substitutions per site. CCR, cation-conducting channelrhodopsin; ACR, anion-conducting channelrhodopsin. An overview of ChRs used to generate the phylogenetic tree can be found in the Supplementary Data 1. b Distribution and relative abundance of MerMAIDs in samples from the Tara Oceans project. Area of each circle indicates the estimated average abundance of MerMAID-like rhodopsins at different Tara Oceans stations. Stations were MerMAIDs were not detected (n.d.) are indicated by crosses. c Photocurrent traces of representative members of previously identified ChR families and MerMAIDs, recorded from −60 to +40 mV in steps of 20 or 15 mV (GtCCR4). Gray bars indicate light application at denoted wavelengths. d, e Desensitization (d) and peak current amplitudes (e) of all MerMAIDs at −60 mV during continuous illumination with 500 nm light. f Normalized action spectrum of MerMAID1. Single measurements are shown as dots (n = 4), and the solid line represents fitted data. Dashed lines indicate light penetration depth in coastal and open seawater (adopted from ref. 93). λmax, maximum response wavelength; g λmax for all MerMAIDs. Mean values (thick lines) ± standard deviation (whiskers) are shown, and single-measurement data points are represented as dots. Source data are provided as a Source Data file (d–g)